Compaction of single DNA molecules induced by binding of integration host factor (IHF)
- PMID: 11535804
- PMCID: PMC58522
- DOI: 10.1073/pnas.181029198
Compaction of single DNA molecules induced by binding of integration host factor (IHF)
Abstract
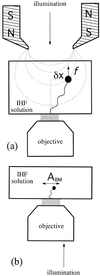
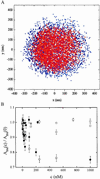
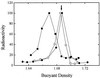
We studied the interaction between the integration host factor (IHF), a major nucleoid-associated protein in bacteria, and single DNA molecules. Force-extension measurements of lambda DNA and an analysis of the Brownian motion of small beads tethered to a surface by single short DNA molecules, in equilibrium with an IHF solution, indicate that: (i) the DNA-IHF complex retains a random, although more compact, coiled configuration for zero or small values of the tension, (ii) IHF induces DNA compaction by binding to multiple DNA sites with low specificity, and (iii) with increasing tension on the DNA, the elastic properties of bare DNA are recovered. This behavior is consistent with the predictions of a statistical mechanical model describing how proteins bending DNA are driven off by an applied tension on the DNA molecule. Estimates of the amount of bound IHF in DNA-IHF complexes obtained from the model agree very well with independent measurements of this quantity obtained from the analysis of DNA-IHF crosslinking. Our findings support the long-held view that IHF and other histone-like proteins play an important role in shaping the long-scale structure of the bacterial nucleoid.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

