Cloning and characterization of a histone deacetylase, HDAC9
- PMID: 11535832
- PMCID: PMC58507
- DOI: 10.1073/pnas.191375098
Cloning and characterization of a histone deacetylase, HDAC9
Abstract
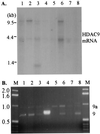
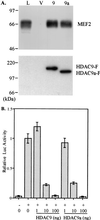
Histone deacetylase (HDAC) catalyzes the removal of the acetyl group from the lysine residues in the N-terminal tails of nucleosomal core histones. Eight human HDACs have been identified so far. Here, we report the identification of a ninth member of the HDAC family, designated HDAC9. HDAC9 is a class II HDAC and its gene resides on human chromosome 7. HDAC9 has several alternatively spliced isoforms. One of these isoforms is histone deacetylase-related protein or myocyte enhancer-binding factor 2-interacting transcriptional repressor that we and others have previously reported and which does not possess an HDAC catalytic domain. The longest of the HDAC9 isoforms contains 1,011 aa. The isoform, designated HDAC9a, is 132 aa shorter at the C terminus than HDAC9. Also, we have identified isoforms of HDAC9 that lack the nuclear localization signal. Similar to histone deacetylase-related protein, HDAC9 transcripts are expressed at high levels in brain and skeletal muscle. The ratio of HDAC9 and HDAC9a transcripts differs among the tissues examined. HDAC9 and HDAC9a contain the HDAC catalytic domain, and Flag-tagged HDAC9 and HDAC9a possess deacetylase activity. HDAC9 and HDAC9a also repress myocyte enhancer-binding factor 2-mediated transcription. In the present study, we have identified HDAC9 and a number of alternatively spliced isoforms of HDAC9 with potentially different biological activities.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

