Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action
- PMID: 11544185
- PMCID: PMC312774
- DOI: 10.1101/gad.205501
Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action
Abstract
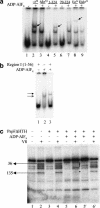
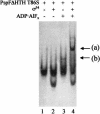
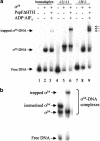
Conformational changes in sigma 54 (sigma(54)) and sigma(54)-holoenzyme depend on nucleotide hydrolysis by an activator. We now show that sigma(54) and its holoenzyme bind to the central ATP-hydrolyzing domains of the transcriptional activators PspF and NifA in the presence of ADP-aluminum fluoride, an analog of ATP in the transition state for hydrolysis. Direct binding of sigma(54) Region I to activator in the presence of ADP-aluminum fluoride was shown and inferred from in vivo suppression genetics. Energy transduction appears to occur through activator contacts to sigma(54) Region I. ADP-aluminum fluoride-dependent interactions and consideration of other AAA+ proteins provide insight into activator mechanochemical action.
Figures
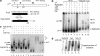


References
-
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. - PubMed
-
- Cannon W, Missailidis S, Smith C, Cottier A, Austin S, Moore M, Buck M. Core RNA polymerase and promoter DNA interactions of purified domains of ςN: Bipartite functions. J Mol Biol. 1995;248:781–803. - PubMed
-
- Cannon WV, Gallegos MT, Buck M. Isomerization of a binary ς–promoter DNA complex by transcription activators. Nat Struct Biol. 2000;7:594–601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
