Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside
- PMID: 11544196
- PMCID: PMC311128
- DOI: 10.1101/gr.164201
Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside
Abstract
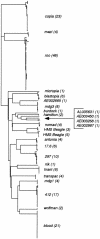
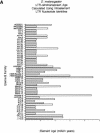
The recent release of the complete euchromatic genome sequence of Drosophila melanogaster offers a unique opportunity to explore the evolutionary history of transposable elements (TEs) within the genome of a higher eukaryote. In this report, we describe the annotation and phylogenetic comparison of 178 full-length long terminal repeat (LTR) retrotransposons from the sequenced component of the D. melanogaster genome. We report the characterization of 17 LTR retrotransposon families described previously and five newly discovered element families. Phylogenetically, these families can be divided into three distinct lineages that consist of members from the canonical Copia and Gypsy groups as well as a newly discovered third group containing BEL, mazi, and roo elements. Each family consists of members with average pairwise identities > or =99% at the nucleotide level, indicating they may be the products of recent transposition events. Consistent with the recent transposition hypothesis, we found that 70% (125/178) of the elements (across all families) have identical intra-element LTRs. Using the synonymous substitution rate that has been calculated previously for Drosophila (.016 substitutions per site per million years) and the intra-element LTR divergence calculated here, the average age of the remaining 30% (53/178) of the elements was found to be 137,000 +/-89,000 yr. Collectively, these results indicate that many full-length LTR retrotransposons present in the D. melanogaster genome have transposed well after this species diverged from its closest relative Drosophila simulans, 2.3 +/-.3 million years ago.
Figures






References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Baldrich E, Dimitri P, Desset S, Leblanc P, Codipietro D, Vaury C. Genomic distribution of the retrovirus-like element ZAM in Drosophila. Genetica. 1997;100:131–140. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
