Assembly of the working draft of the human genome with GigAssembler
- PMID: 11544197
- PMCID: PMC311095
- DOI: 10.1101/gr.183201
Assembly of the working draft of the human genome with GigAssembler
Abstract
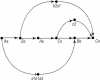
The data for the public working draft of the human genome contains roughly 400,000 initial sequence contigs in approximately 30,000 large insert clones. Many of these initial sequence contigs overlap. A program, GigAssembler, was built to merge them and to order and orient the resulting larger sequence contigs based on mRNA, paired plasmid ends, EST, BAC end pairs, and other information. This program produced the first publicly available assembly of the human genome, a working draft containing roughly 2.7 billion base pairs and covering an estimated 88% of the genome that has been used for several recent studies of the genome. Here we describe the algorithm used by GigAssembler.
Figures



Comment in
-
Assembling puzzles from preassembled blocks.Genome Res. 2001 Sep;11(9):1461-2. doi: 10.1101/gr.206301. Genome Res. 2001. PMID: 11544188 No abstract available.
References
-
- Anson E, Myers G. Proc. RECOMB '99, Lyon, France. 1999. Algorithms for whole genome shotgun sequencing; pp. 1–9.
-
- Bentley DR, Deloukas P, Dunham A, French L, Gregory SG, Humphrey SJ, Mungall AJ, Ross MT, Carter NP, Dunham I, et al. The physical maps for sequencing human chromosomes 1, 6, 9, 10, 13, 20 and X. Nature. 2001;409:942–943. - PubMed
-
- Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
