A predictive model for regulatory sequences directing liver-specific transcription
- PMID: 11544200
- PMCID: PMC311083
- DOI: 10.1101/gr.180601
A predictive model for regulatory sequences directing liver-specific transcription
Abstract
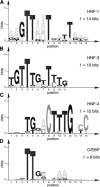
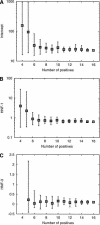
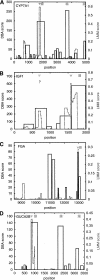
The identification and interpretation of the regulatory signals within the human genome remain among the greatest goals and most difficult challenges in genome analysis. The ability to predict the temporal and spatial control of transcription is likely to require a combination of methods to address the contribution of sequence-specific signals, protein-protein interactions and chromatin structure. We present here a new procedure to identify clusters of transcription factor binding sites characteristic of sequence modules experimentally verified to direct transcription selectively to liver cells. This algorithm is sufficiently specific to identify known regulatory sequences in genes selectively expressed in liver, promising acceleration of experimental promoter analysis. In combination with phylogenetic footprinting, this improvement in the specificity of predictions is sufficient to motivate a scan of the human genome. Potential regulatory modules were identified in orthologous human and rodent genomic sequences containing both known and uncharacterized genes.
Figures



References
-
- Bernard P, Goudonnet H, Artur Y, Desvergne B, Wahli W. Activation of the mouse TATA-less and human TATA-containing UDP-glucuronosyltransferase 1A1 promoters by hepatocyte nuclear factor 1. Mol Pharmacol. 1999;56:526–536. - PubMed
-
- Blackwood EM, Kadonaga JT. Going the distance: A current view of enhancer action. Science. 1998;281:61–63. - PubMed
-
- Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;97:267–282. - PubMed
-
- Claverie JM. From bioinformatics to computational biology. Genome Res. 2000;10:1277–1279. - PubMed
-
- Cooper AD, Chen J, Botelho-Yetkinler MJ, Cao Y, Taniguchi T, Levy-Wilson B. Characterization of hepatic-specific regulatory elements in the promoter region of the human cholesterol 7α-hydroxylase gene. J Biol Chem. 1997;272:3444–3452. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
