cis element/transcription factor analysis (cis/TF): a method for discovering transcription factor/cis element relationships
- PMID: 11544201
- PMCID: PMC311103
- DOI: 10.1101/gr.158301
cis element/transcription factor analysis (cis/TF): a method for discovering transcription factor/cis element relationships
Abstract
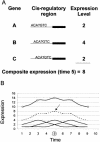
We report a simple new algorithm, cis/TF, that uses genomewide expression data and the full genomic sequence to match transcription factors to their binding sites. Most previous computational methods discovered binding sites by clustering genes having similar expression patterns and then identifying over-represented subsequences in the promoter regions of those genes. By contrast, cis/TF asserts that B is a likely binding site of a transcription factor T if the expression pattern of T is correlated to the composite expression patterns of all genes containing B, even when those genes are not mutually correlated. Thus, our method focuses on binding sites rather than genes. The algorithm has successfully identified experimentally-supported transcription factor binding relationships in tests on several data sets from Saccharomyces cerevisiae.
Figures


References
-
- Albrecht G, Mösch HU, Hoffmann B, Reusser U, Braus GH. Monitoring the GCN4 protein-mediated response in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:12696–12702. - PubMed
-
- Arnone MI, Davidson EH. The hardwiring of development: Organization and function of genomic regulatory systems. Development. 1997;128:1851–1864. - PubMed
-
- Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with genome-wide mRNA expression data. Nat Genet. 2001;27:167–171. - PubMed
-
- Cherry, J.M., Ball, C., Dolinski, K., Dwight, S., Harris, M., Matese, J.C., Sherlock, G., Binkley, G., Jin, H., Weng, S., et al. Saccharomyces Genome Database http://genome-ftp.stanford.edu/pub/yeast/SacchDB/. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
