Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a
- PMID: 11544215
- PMCID: PMC95444
- DOI: 10.1128/JB.183.19.5535-5543.2001
Nested deletions of the SRL pathogenicity island of Shigella flexneri 2a
Abstract
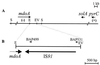
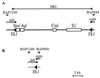
In this study, we determined the boundaries of a 99-kb deletable element of Shigella flexneri 2a strain YSH6000. The element, designated the multiple-antibiotic resistance deletable element (MRDE), had recently been found to contain a 66-kb pathogenicity island (PAI)-like element (designated the SRL PAI) which carries the Shigella resistance locus (SRL), encoding resistance determinants to streptomycin, ampicillin, chloramphenicol, and tetracycline. The YSH6000 MRDE was found to be flanked by two identical IS91 elements present at the S. flexneri homologs of the Escherichia coli genes putA and mdoA on NotI fragment D. Sequence data from two YSH6000-derived MRDE deletants, YSH6000T and S2430, revealed that deletion of the MRDE occurred between the two flanking IS91 elements, resulting in a single IS91 element spanning the two original IS91 loci. Selection for the loss of tetracycline resistance confirmed that the MRDE deletion occurred reproducibly from the same chromosomal site and also showed that the SRL PAI and the SRL itself were capable of independent deletion from the chromosome, thus revealing a unique set of nested deletions. The excision frequency of the SRL PAI was estimated to be 10(-5) per cell in the wild type, and mutation of a P4-like integrase gene (int) at the left end of the SRL PAI revealed that int mediates precise deletion of the PAI.
Figures



Similar articles
-
Genetic organization of the she pathogenicity island in Shigella flexneri 2a.Microb Pathog. 2001 Jan;30(1):1-8. doi: 10.1006/mpat.2000.0404. Microb Pathog. 2001. PMID: 11162180
-
Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes.Infect Immun. 2001 Oct;69(10):6012-21. doi: 10.1128/IAI.69.10.6012-6021.2001. Infect Immun. 2001. PMID: 11553538 Free PMC article.
-
Regulated site-specific recombination of the she pathogenicity island of Shigella flexneri.Mol Microbiol. 2004 Jun;52(5):1329-36. doi: 10.1111/j.1365-2958.2004.04048.x. Mol Microbiol. 2004. PMID: 15165236
-
Pathogenicity island integrase cross-talk: a potential new tool for virulence modulation.Mol Microbiol. 2006 Aug;61(3):555-9. doi: 10.1111/j.1365-2958.2006.05262.x. Mol Microbiol. 2006. PMID: 16879637 Review.
-
A chromosome map of Shigella flexneri with the loci related to pathogenicity.Mol Gen Mikrobiol Virusol. 1994 Jan-Feb;(1):3-10. Mol Gen Mikrobiol Virusol. 1994. PMID: 8133848 Review. No abstract available.
Cited by
-
Molecular epidemiology of the SRL pathogenicity island.Antimicrob Agents Chemother. 2003 Feb;47(2):727-34. doi: 10.1128/AAC.47.2.727-734.2003. Antimicrob Agents Chemother. 2003. PMID: 12543684 Free PMC article.
-
Genome Analysis of Shigella flexneri Serotype 3b Strain SFL1520 Reveals Significant Horizontal Gene Acquisitions Including a Multidrug Resistance Cassette.Genome Biol Evol. 2019 Mar 1;11(3):776-785. doi: 10.1093/gbe/evz026. Genome Biol Evol. 2019. PMID: 30715343 Free PMC article.
-
Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar Typhi.J Bacteriol. 2004 May;186(10):3202-13. doi: 10.1128/JB.186.10.3202-3213.2004. J Bacteriol. 2004. PMID: 15126483 Free PMC article.
-
SRL pathogenicity island contributes to the metabolism of D-aspartate via an aspartate racemase in Shigella flexneri YSH6000.PLoS One. 2020 Jan 24;15(1):e0228178. doi: 10.1371/journal.pone.0228178. eCollection 2020. PLoS One. 2020. PMID: 31978153 Free PMC article.
-
Instability of pathogenicity islands in uropathogenic Escherichia coli 536.J Bacteriol. 2004 May;186(10):3086-96. doi: 10.1128/JB.186.10.3086-3096.2004. J Bacteriol. 2004. PMID: 15126470 Free PMC article.
References
-
- Al-Hasani K, Rajakumar K, Bulach D, Robins-Browne R, Adler B, Sakellaris H. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb Pathog. 2001;30:1–8. - PubMed
-
- Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1993.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

