Genetics and regulation of chitobiose utilization in Borrelia burgdorferi
- PMID: 11544216
- PMCID: PMC95445
- DOI: 10.1128/JB.183.19.5544-5553.2001
Genetics and regulation of chitobiose utilization in Borrelia burgdorferi
Abstract
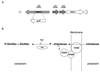
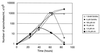
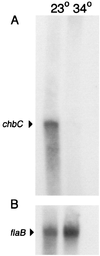
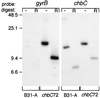
Borrelia burgdorferi spends a significant proportion of its life cycle within an ixodid tick, which has a cuticle containing chitin, a polymer of N-acetylglucosamine (GlcNAc). The B. burgdorferi celA, celB, and celC genes encode products homologous to transporters for cellobiose and chitobiose (the dimer subunit of chitin) in other bacteria, which could be useful for bacterial nutrient acquisition during growth within ticks. We found that chitobiose efficiently substituted for GlcNAc during bacterial growth in culture medium. We inactivated the celB gene, which encodes the putative membrane-spanning component of the transporter, and compared growth of the mutant in various media to that of its isogenic parent. The mutant was no longer able to utilize chitobiose, while neither the mutant nor the wild type can utilize cellobiose. We propose renaming the three genes chbA, chbB, and chbC, since they probably encode a chitobiose transporter. We also found that the chbC gene was regulated in response to growth temperature and during growth in medium lacking GlcNAc.
Figures





Similar articles
-
The chitobiose transporter, chbC, is required for chitin utilization in Borrelia burgdorferi.BMC Microbiol. 2010 Jan 26;10:21. doi: 10.1186/1471-2180-10-21. BMC Microbiol. 2010. PMID: 20102636 Free PMC article.
-
Chitobiose utilization in Borrelia burgdorferi is dually regulated by RpoD and RpoS.BMC Microbiol. 2009 May 27;9:108. doi: 10.1186/1471-2180-9-108. BMC Microbiol. 2009. PMID: 19473539 Free PMC article.
-
Infectious cycle analysis of a Borrelia burgdorferi mutant defective in transport of chitobiose, a tick cuticle component.Vector Borne Zoonotic Dis. 2004 Summer;4(2):159-68. doi: 10.1089/1530366041210738. Vector Borne Zoonotic Dis. 2004. PMID: 15228817
-
Pathophysiology of the Lyme disease spirochete, Borrelia burgdorferi, in ixodid ticks.Rev Infect Dis. 1989 Sep-Oct;11 Suppl 6:S1442-50. doi: 10.1093/clinids/11.supplement_6.s1442. Rev Infect Dis. 1989. PMID: 2682956 Review.
-
Epizootiology of Lyme disease and methods of cultivating Borrelia burgdorferi.Ann N Y Acad Sci. 1992 Jun 16;653:52-63. doi: 10.1111/j.1749-6632.1992.tb19629.x. Ann N Y Acad Sci. 1992. PMID: 1626892 Review.
Cited by
-
Interaction of the Lyme disease spirochete with its tick vector.Cell Microbiol. 2016 Jul;18(7):919-27. doi: 10.1111/cmi.12609. Epub 2016 May 24. Cell Microbiol. 2016. PMID: 27147446 Free PMC article. Review.
-
Study of the response regulator Rrp1 reveals its regulatory role in chitobiose utilization and virulence of Borrelia burgdorferi.Infect Immun. 2013 May;81(5):1775-87. doi: 10.1128/IAI.00050-13. Epub 2013 Mar 11. Infect Immun. 2013. PMID: 23478317 Free PMC article.
-
The chitobiose transporter, chbC, is required for chitin utilization in Borrelia burgdorferi.BMC Microbiol. 2010 Jan 26;10:21. doi: 10.1186/1471-2180-10-21. BMC Microbiol. 2010. PMID: 20102636 Free PMC article.
-
Characterization of the RelBbu Regulon in Borrelia burgdorferi Reveals Modulation of Glycerol Metabolism by (p)ppGpp.PLoS One. 2015 Feb 17;10(2):e0118063. doi: 10.1371/journal.pone.0118063. eCollection 2015. PLoS One. 2015. PMID: 25688856 Free PMC article.
-
Differential telomere processing by Borrelia telomere resolvases in vitro but not in vivo.J Bacteriol. 2006 Nov;188(21):7378-86. doi: 10.1128/JB.00760-06. Epub 2006 Aug 25. J Bacteriol. 2006. PMID: 16936037 Free PMC article.
References
-
- Bono J L, Tilly K, Stevenson B, Hogan D, Rosa P. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology. 1998;144:1033–1044. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

