Analysis of functional domains of the Enterococcus faecalis pheromone-induced surface protein aggregation substance
- PMID: 11544229
- PMCID: PMC95458
- DOI: 10.1128/JB.183.19.5659-5667.2001
Analysis of functional domains of the Enterococcus faecalis pheromone-induced surface protein aggregation substance
Abstract
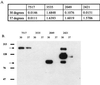
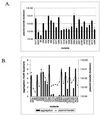
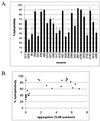
Pheromone-inducible aggregation substance (AS) proteins of Enterococcus faecalis are essential for high-efficiency conjugation of the sex pheromone plasmids and also serve as virulence factors during host infection. A number of different functions have been attributed to AS in addition to bacterial cell aggregation, including adhesion to host cells, adhesion to fibrin, increased cell surface hydrophobicity, resistance to killing by polymorphonuclear leukocytes and macrophages, and increased vegetation size in an experimental endocarditis model. Relatively little information is available regarding the structure-activity relationship of AS. To identify functional domains, a library of 23 nonpolar 31-amino-acid insertions was constructed in Asc10, the AS encoded by the plasmid pCF10, using the transposons TnlacZ/in and TnphoA/in. Analysis of these insertions revealed a domain necessary for donor-recipient aggregation that extends further into the amino terminus of the protein than previously reported. In addition, insertions in the C terminus of the protein also reduced aggregation. As expected, the ability to aggregate correlates with efficient plasmid transfer. The results also indicated that an increase in cell surface hydrophobicity resulting from AS expression is not sufficient to mediate bacterial aggregation.
Figures




Similar articles
-
Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis.J Bacteriol. 1991 Dec;173(23):7650-64. doi: 10.1128/jb.173.23.7650-7664.1991. J Bacteriol. 1991. PMID: 1938961 Free PMC article.
-
Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence.Infect Immun. 2009 Jan;77(1):539-48. doi: 10.1128/IAI.01034-08. Epub 2008 Oct 27. Infect Immun. 2009. PMID: 18955479 Free PMC article.
-
An amino-terminal domain of Enterococcus faecalis aggregation substance is required for aggregation, bacterial internalization by epithelial cells and binding to lipoteichoic acid.Mol Microbiol. 2004 May;52(4):1159-71. doi: 10.1111/j.1365-2958.2004.04045.x. Mol Microbiol. 2004. PMID: 15130132
-
The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism?Eur J Biochem. 1994 Jun 1;222(2):235-46. doi: 10.1111/j.1432-1033.1994.tb18862.x. Eur J Biochem. 1994. PMID: 8020463 Review.
-
The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution.Philos Trans R Soc Lond B Biol Sci. 2007 Jul 29;362(1483):1185-93. doi: 10.1098/rstb.2007.2043. Philos Trans R Soc Lond B Biol Sci. 2007. PMID: 17360276 Free PMC article. Review.
Cited by
-
Functional analysis of TraA, the sex pheromone receptor encoded by pPD1, in a promoter region essential for the mating response in Enterococcus faecalis.J Bacteriol. 2002 Nov;184(22):6343-50. doi: 10.1128/JB.184.22.6343-6350.2002. J Bacteriol. 2002. PMID: 12399504 Free PMC article.
-
Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence.Mol Microbiol. 2015 Feb;95(4):660-77. doi: 10.1111/mmi.12893. Epub 2014 Dec 30. Mol Microbiol. 2015. PMID: 25431047 Free PMC article.
-
A sampling survey of enterococci within pasteurized, fermented dairy products and their virulence and antibiotic resistance properties.PLoS One. 2021 Jul 15;16(7):e0254390. doi: 10.1371/journal.pone.0254390. eCollection 2021. PLoS One. 2021. PMID: 34264984 Free PMC article.
-
Insights into ecology, pathogenesis, and biofilm formation of Enterococcus faecalis from functional genomics.Microbiol Mol Biol Rev. 2025 Mar 27;89(1):e0008123. doi: 10.1128/mmbr.00081-23. Epub 2024 Dec 23. Microbiol Mol Biol Rev. 2025. PMID: 39714182 Review.
-
The Tra domain of the lactococcal CluA surface protein is a unique domain that contributes to sex factor DNA transfer.J Bacteriol. 2006 Mar;188(6):2106-14. doi: 10.1128/JB.188.6.2106-2114.2006. J Bacteriol. 2006. PMID: 16513740 Free PMC article.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990.
-
- Bae T, Clerc-Bardin S, Dunny G M. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J Mol Biol. 2000;297:861–875. - PubMed
-
- Bryan E M, Bae T, Kleerebezem H, Dunny G M. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid. 2000;44:183–190. - PubMed
-
- Christie P J, Dunny G M. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid. 1986;15:230–241. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

