Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP
- PMID: 11544232
- PMCID: PMC95461
- DOI: 10.1128/JB.183.19.5684-5697.2001
Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP
Abstract
The complete 108,845-nucleotide sequence of catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP was determined. Plasmid pADP-1 was previously shown to encode AtzA, AtzB, and AtzC, which catalyze the sequential hydrolytic removal of s-triazine ring substituents from the herbicide atrazine to yield cyanuric acid. Computational analyses indicated that pADP-1 encodes 104 putative open reading frames (ORFs), which are predicted to function in catabolism, transposition, and plasmid maintenance, transfer, and replication. Regions encoding transfer and replication functions of pADP-1 had 80 to 100% amino acid sequence identity to pR751, an IncPbeta plasmid previously isolated from Enterobacter aerogenes. pADP-1 was shown to contain a functional mercury resistance operon with 99% identity to Tn5053. Complete copies of transposases with 99% amino acid sequence identity to TnpA from IS1071 and TnpA from Pseudomonas pseudoalcaligenes were identified and flank each of the atzA, atzB, and atzC genes, forming structures resembling nested catabolic transposons. Functional analyses identified three new catabolic genes, atzD, atzE, and atzF, which participate in atrazine catabolism. Crude extracts from Escherichia coli expressing AtzD hydrolyzed cyanuric acid to biuret. AtzD showed 58% amino acid sequence identity to TrzD, a cyanuric acid amidohydrolase, from Pseudomonas sp. strain NRRLB-12227. Two other genes encoding the further catabolism of cyanuric acid, atzE and atzF, reside in a contiguous cluster adjacent to a potential LysR-type transcriptional regulator. E. coli strains bearing atzE and atzF were shown to encode a biuret hydrolase and allophanate hydrolase, respectively. atzDEF are cotranscribed. AtzE and AtzF are members of a common amidase protein family. These data reveal the complete structure of a catabolic plasmid and show that the atrazine catabolic genes are dispersed on three disparate regions of the plasmid. These results begin to provide insight into how plasmids are structured, and thus evolve, to encode the catabolism of compounds recently added to the biosphere.
Figures
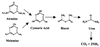





References
-
- Aramaki H, Yagi N, Suzuki M. Residues important for the function of a multihelical DNA binding domain in the new transcription factor family of Cam and Tet repressors. Protein Eng. 1995;8:1259–1266. - PubMed
-
- Austen R A, Dunn N W. Isolation of mutants with altered metabolic control of the NAH plasmid-encoded catechol meta-cleavage pathway. Aust J Biol Sci. 1977;30:583–592. - PubMed
-
- Bagdasarian M, Lurz R, Ruckert B, Franklin F C, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

