Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15
- PMID: 11549765
- PMCID: PMC139453
- DOI: 10.1105/tpc.010113
Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15
Abstract
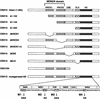
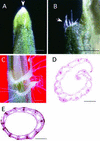
The rice KNOX protein OSH15 consists of four conserved domains: the MEINOX domain, which can be divided into two subdomains (KNOX1 and KNOX2); the GSE domain; the ELK domain; and the homeodomain (HD). To investigate the function of each domain, we generated 10 truncated proteins with deletions in the conserved domains and four proteins with mutations in the conserved amino acids in the HD. Transgenic analysis suggested that KNOX2 and HD are essential for inducing the abnormal phenotype and that the KNOX1 and ELK domains affect phenotype severity. We also found that both KNOX2 and HD are necessary for homodimerization and that only HD is needed for binding of OSH15 to its target sequence. Transactivation studies suggested that both the KNOX1 and ELK domains play a role in suppressing target gene expression. On the basis of these findings, we propose that overproduced OSH15 probably acts as a dimer and may ectopically suppress the expression of target genes that induce abnormal morphology in transgenic plants.
Figures








References
-
- Affolter, M., Schier, A., and Gehring, W.J. (1990). Homeodomain proteins and regulation of gene expression. Curr. Opin. Cell Biol. 2 485–495. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

