The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed
- PMID: 11549766
- PMCID: PMC139454
- DOI: 10.1105/tpc.010098
The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed
Abstract
In Arabidopsis, proanthocyanidins specifically accumulate in the endothelium during early seed development. At least three TRANSPARENT TESTA (TT) genes, TT2, TT8, and TTG1, are necessary for the normal expression of several flavonoid structural genes in immature seed, such as DIHYDROFLAVONOL-4-REDUCTASE and BANYULS (BAN). TT8 and TTG1 were characterized recently and found to code for a basic helix-loop-helix domain transcription factor and a WD-repeat-containing protein, respectively. Here the molecular cloning of the TT2 gene was achieved by T-DNA tagging. TT2 encoded an R2R3 MYB domain protein with high similarity to the rice OsMYB3 protein and the maize COLORLESS1 factor. A TT2-green fluorescent protein fusion protein was located mostly in the nucleus, in agreement with the regulatory function of the native TT2 protein. TT2 expression was restricted to the seed during early embryogenesis, consistent with BAN expression and the proanthocyanidin deposition profile. Finally, in gain-of-function experiments, TT2 was able to induce ectopic expression of BAN in young seedlings and roots in the presence of a functional TT8 protein. Therefore, our results strongly suggest that stringent spatial and temporal BAN expression, and thus proanthocyanidin accumulation, are determined at least partially by TT2.
Figures

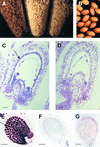
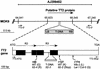




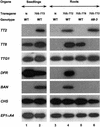
References
-
- Aastrup, S., Outtrup, H., and Erdal, K. (1984). Location of the proanthocyanidins in the barley grain. Carlsberg Res. Commun. 49 105–109.
-
- Albert, S., Delseny, M., and Devic, M. (1997). BANYULS, a novel negative regulator of flavonoid biosynthesis in the Arabidopsis seed coat. Plant J. 11 289–299. - PubMed
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316 1194–1199.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

