Distinct regions in the 3' untranslated region are responsible for targeting and stabilizing utrophin transcripts in skeletal muscle cells
- PMID: 11551978
- PMCID: PMC2150820
- DOI: 10.1083/jcb.200101108
Distinct regions in the 3' untranslated region are responsible for targeting and stabilizing utrophin transcripts in skeletal muscle cells
Abstract
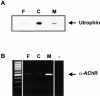
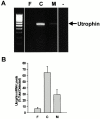
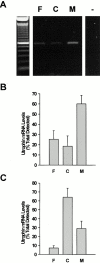
In this study, we have sought to determine whether utrophin transcripts are targeted to a distinct subcellular compartment in skeletal muscle cells, and have examined the role of the 3' untranslated region (UTR) in regulating the stability and localization of utrophin transcripts. Our results show that utrophin transcripts associate preferentially with cytoskeleton-bound polysomes via actin microfilaments. Because this association is not evident in myoblasts, our findings also indicate that the localization of utrophin transcripts with cytoskeleton-bound polysomes is under developmental influences. Transfection of LacZ reporter constructs containing the utrophin 3'UTR showed that this region is critical for targeting chimeric mRNAs to cytoskeleton-bound polysomes and controlling transcript stability. Deletion studies resulted in the identification of distinct regions within the 3'UTR responsible for targeting and stabilizing utrophin mRNAs. Together, these results illustrate the contribution of posttranscriptional events in the regulation of utrophin in skeletal muscle. Accordingly, these findings provide novel targets, in addition to transcriptional events, for which pharmacological interventions may be envisaged to ultimately increase the endogenous levels of utrophin in skeletal muscle fibers from Duchenne muscular dystrophy (DMD) patients.
Figures





Similar articles
-
Multiple regulatory events controlling the expression and localization of utrophin in skeletal muscle fibers: insights into a therapeutic strategy for Duchenne muscular dystrophy.J Physiol Paris. 2002 Jan-Mar;96(1-2):31-42. doi: 10.1016/s0928-4257(01)00078-x. J Physiol Paris. 2002. PMID: 11755781 Review.
-
Increased expression of utrophin in a slow vs. a fast muscle involves posttranscriptional events.Am J Physiol Cell Physiol. 2001 Oct;281(4):C1300-9. doi: 10.1152/ajpcell.2001.281.4.C1300. Am J Physiol Cell Physiol. 2001. PMID: 11546668
-
The role of basal and myogenic factors in the transcriptional activation of utrophin promoter A: implications for therapeutic up-regulation in Duchenne muscular dystrophy.Nucleic Acids Res. 2001 Dec 1;29(23):4843-50. doi: 10.1093/nar/29.23.4843. Nucleic Acids Res. 2001. PMID: 11726694 Free PMC article.
-
Activation of p38 signaling increases utrophin A expression in skeletal muscle via the RNA-binding protein KSRP and inhibition of AU-rich element-mediated mRNA decay: implications for novel DMD therapeutics.Hum Mol Genet. 2013 Aug 1;22(15):3093-111. doi: 10.1093/hmg/ddt165. Epub 2013 Apr 10. Hum Mol Genet. 2013. PMID: 23575223
-
Regulation and functional significance of utrophin expression at the mammalian neuromuscular synapse.Microsc Res Tech. 2000 Apr 1;49(1):90-100. doi: 10.1002/(SICI)1097-0029(20000401)49:1<90::AID-JEMT10>3.0.CO;2-L. Microsc Res Tech. 2000. PMID: 10757882 Review.
Cited by
-
Viral-mediated gene therapy for the muscular dystrophies: successes, limitations and recent advances.Biochim Biophys Acta. 2007 Feb;1772(2):243-62. doi: 10.1016/j.bbadis.2006.09.007. Epub 2006 Sep 26. Biochim Biophys Acta. 2007. PMID: 17064882 Free PMC article. Review.
-
Biglycan recruits utrophin to the sarcolemma and counters dystrophic pathology in mdx mice.Proc Natl Acad Sci U S A. 2011 Jan 11;108(2):762-7. doi: 10.1073/pnas.1013067108. Epub 2010 Dec 27. Proc Natl Acad Sci U S A. 2011. PMID: 21187385 Free PMC article.
-
Conserved regions of the DMD 3' UTR regulate translation and mRNA abundance in cultured myotubes.Neuromuscul Disord. 2014 Aug;24(8):693-706. doi: 10.1016/j.nmd.2014.05.006. Epub 2014 May 22. Neuromuscul Disord. 2014. PMID: 24928536 Free PMC article.
-
Modulation of utrophin A mRNA stability in fast versus slow muscles via an AU-rich element and calcineurin signaling.Nucleic Acids Res. 2008 Feb;36(3):826-38. doi: 10.1093/nar/gkm1107. Epub 2007 Dec 15. Nucleic Acids Res. 2008. PMID: 18084024 Free PMC article.
-
Converging pathways involving microRNA-206 and the RNA-binding protein KSRP control post-transcriptionally utrophin A expression in skeletal muscle.Nucleic Acids Res. 2014 Apr;42(6):3982-97. doi: 10.1093/nar/gkt1350. Epub 2013 Dec 26. Nucleic Acids Res. 2014. PMID: 24371285 Free PMC article.
References
-
- Adamou, J., and J. Bag. 1992. Alteration of translation and stability of mRNA for the poly(A)-binding protein during myogenesis. Eur. J. Biochem. 209:803–812. - PubMed
-
- Ahn, A.H., and L.M. Kunkel. 1993. The structural and functional diversity of dystrophin. Nat. Genet. 3:283–291. - PubMed
-
- Bassell, G.J., and R.H. Singer. 1997. mRNA and cytoskeletal filaments. Curr. Opin. Cell Biol. 9:109–115. - PubMed
-
- Bassell, G.J., R.H. Singer, and K.S. Kosik. 1994. Association of Poly(A) mRNA with microtubules in cultured neurons. Neuron. 12:571–582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

