Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage
- PMID: 11553559
- PMCID: PMC98750
- DOI: 10.1128/IAI.69.10.6186-6192.2001
Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage
Abstract
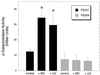
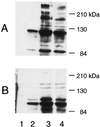
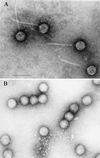
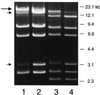
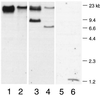
The binding of platelets by bacteria is a proposed central mechanism in the pathogenesis of infective endocarditis. Platelet binding by Streptococcus mitis strain SF100 (an endocarditis isolate) was recently shown to be mediated in part by the surface proteins PblA and PblB. The genes encoding PblA and PblB are clustered with genes nearly identical to those of streptococcal phages r1t, 01205, and Dp-1, suggesting that pblA and pblB might reside within a prophage. To address this possibility, cultures of SF100 were exposed to either mitomycin C or UV light, both of which are known to induce the lytic cycle of many temperate phages. Both treatments caused a significant increase in the transcription of pblA. Treatment with mitomycin C or UV light also caused a substantial increase in the expression of PblA and PblB, as detected by Western blot analysis of proteins in the SF100 cell wall. By electron microscopy, phage particles were readily visible in the supernatants from induced cultures of SF100. The phage, designated SM1, had a double-stranded DNA genome of approximately 35 kb. Southern blot analysis of phage DNA indicated that pblA and pblB were contained within the SM1 genome. Furthermore, Western blot analysis of phage proteins revealed that both PblA and PblB were present in the phage particles. These findings indicate that PblA and PblB are encoded by a lysogenic bacteriophage, which could facilitate the dissemination of these potential virulence determinants to other bacterial pathogens.
Figures

References
-
- Barondess J J, Beckwith J. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature. 1990;346:871–874. - PubMed
-
- Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol Microbiol. 1997;25:285–294. - PubMed
-
- Claverys J P, Dintilhac A, Pestova E V, Martin B, Morrison D A. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–128. - PubMed
-
- Coleman D C, Sullivan D J, Russell R J, Arbuthnott J P, Carey B F, Pomeroy H M. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J Gen Microbiol. 1989;135:1679–1697. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

