Use of operon fusions in Mannheimia haemolytica to identify environmental and cis-acting regulators of leukotoxin transcription
- PMID: 11553565
- PMCID: PMC98756
- DOI: 10.1128/IAI.69.10.6231-6239.2001
Use of operon fusions in Mannheimia haemolytica to identify environmental and cis-acting regulators of leukotoxin transcription
Abstract
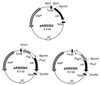
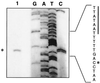
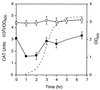
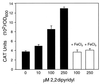
The leukotoxin of Mannheimia haemolytica is an important virulence factor that contributes to much of the pathology observed in the lungs of animals with bovine shipping fever pneumonia. We believe that identification of factors that regulate leukotoxin expression may provide insight into M. haemolytica pathogenicity. The DNA sequence upstream of the leukotoxin operon is divergently shared by P(lapT), which transcribes an arginine permease gene. The intergenic region contains several elements that are potential sites for transcriptional modulation of the promoters. We have developed plasmid-borne chloramphenicol acetyltransferase (cat) operon fusions, as well as lktC::cat chromosomal fusions, to study transcription initiation in M. haemolytica. Using these genetic tools, we have identified cis-acting sequences and environmental conditions that modulate transcription of the leukotoxin and lapT promoters. By deletion analysis, promoters were shown to rely on sequences upstream of their -10 and -35 regions for full activity. Direct repeats of the sequence TGT-N(11)-ACA and a static bend region caused by phased adenine tracts were necessary for full activation of P(lkt). A computer-generated model of the promoter's structure shows how DNA bending brings the repeat sequences within close proximity to the P(lkt) RNA polymerase, and we hypothesize that these repeats are a binding site for an activator of leukotoxin transcription. The lktC::cat operon fusion was also used to demonstrate that, like that of other RTX toxins, leukotoxin transcription is environmentally regulated. Roles for iron deprivation and temperature change were identified.
Figures



References
-
- Adler H I. The use of microbial membranes to achieve anaerobiosis. Crit Rev Biotechnol. 1990;10(2):119–127. - PubMed
-
- Bailey M J A, Koronakis V, Schmoll T, Hughes C. Escherichia coli HlyT protein, a transcriptional activator of haemolysin synthesis and secretion, is encoded by the rfaH (sfrB) locus required for expression of sex factor and lipopolysaccharide genes. Mol Microbiol. 1992;6:1003–1012. - PubMed
-
- Cannon W, Charlton W, Buck M. Organization and function of binding sites for the transcriptional activator NifA in the Klebsiella pneumoniae nifE and nifU promoters. J Mol Biol. 1991;220:915–931. - PubMed
-
- Caskey L S, Lamphear J G, Highlander S K. Binding-protein-dependent arginine transport in Pasteurella haemolytica. Microbiology. 1996;142:1739–1747. - PubMed
-
- Clinkenbeard K D, Upton M L. Lysis of bovine platelets by Pasteurella haemolytica leukotoxin. Am J Vet Res. 1991;52:453–457. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

