Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens
- PMID: 11553577
- PMCID: PMC98768
- DOI: 10.1128/IAI.69.10.6323-6335.2001
Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens
Abstract
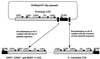
The family of attaching and effacing (A/E) bacterial pathogens, which includes diarrheagenic enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC), remains a significant threat to human and animal health. These bacteria intimately attach to host intestinal cells, causing the effacement of brush border microvilli. The genes responsible for this phenotype are encoded in a pathogenicity island called the locus of enterocyte effacement (LEE). Citrobacter rodentium is the only known murine A/E pathogen and serves as a small animal model for EPEC and EHEC infections. Here we report the full DNA sequence of C. rodentium LEE and provide a comparative analysis with the published LEEs from EPEC, EHEC, and the rabbit diarrheagenic E. coli strain RDEC-1. Although C. rodentium LEE shows high similarities throughout the entire sequence and shares all 41 open reading frames with the LEE from EPEC, EHEC, and RDEC-1, it is unique in its location of the rorf1 and rorf2/espG genes and the presence of several insertion sequences (IS) and IS remnants. The LEE of EPEC and EHEC is inserted into the selC tRNA gene. In contrast, the Citrobacter LEE is flanked on one side by an operon encoding an ABC transport system, and an IS element and sequences homologous to Shigella plasmid R100 and EHEC pO157 flank the other. The presence of plasmid sequences next to C. rodentium LEE suggests that the prototype LEE resided on a horizontally transferable plasmid. Additional sequence analysis reveals that the 3-kb plasmid in C. rodentium is nearly identical to p9705 in EHEC O157:H7, suggesting that horizontal plasmid transfer among A/E pathogens has occurred. Our results indicate that the LEE has been acquired by C. rodentium and A/E E. coli strains independently during evolution.
Figures




References
-
- Barthold S W, Coleman G L, Jacoby R O, Livestone E M, Jonas A M. Transmissible murine colonic hyperplasia. Vet Pathol. 1978;15:223–236. - PubMed
-
- Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

