ASH1 mRNA localization in three acts
- PMID: 11553699
- PMCID: PMC59695
- DOI: 10.1091/mbc.12.9.2567
ASH1 mRNA localization in three acts
Abstract
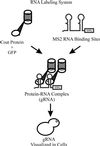
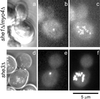
Novel green fluorescent protein (GFP) labeling techniques targeting specific mRNA transcripts reveal discrete phases of mRNA localization in yeast: packaging, transport, and docking. In budding yeast, ASH1 mRNA is translocated via actin and myosin to the tip of growing cells. A GFP-decorated reporter transcript containing the ASH1 3' untranslated region gRNA(ASH1) forms spots of fluorescence localized to a cortical domain at the bud tip, relocates to the mother-bud neck before cell separation, and finally migrates to the incipient bud site before the next budding cycle. The correct positioning of the mRNA requires at least six proteins: She1p-5p and Bud6p/Aip3p. gRNA(ASH1) localization in mutant strains identified three functional categories for the She proteins: mRNA particle formation (She2p and She4p), mRNA transport into the bud (She1p/Myo4p and She3p), and mRNA tethering at the bud tip (She5p/Bni1p and Bud6p/Aip3p). Because localization of the mRNA within the bud does not a priori restrict the translated protein, we examine the distribution of a mother-specific protein (Yta6p) translated from a mRNA directed into the bud. Yta6p remains associated with the mother cortex despite localization of the mRNA to the bud. This video essay traces the life history of a localized mRNA transcript, describes the roles of proteins required to polarize and anchor the mRNA, and demonstrates at least one instance where mRNA localization does not effect protein localization.
Figures





Similar articles
-
Localization and anchoring of mRNA in budding yeast.Curr Biol. 1999 Jun 3;9(11):569-78. doi: 10.1016/s0960-9822(99)80260-7. Curr Biol. 1999. PMID: 10359695
-
She3p possesses a novel activity required for ASH1 mRNA localization in Saccharomyces cerevisiae.Eukaryot Cell. 2009 Jul;8(7):1072-83. doi: 10.1128/EC.00084-09. Epub 2009 May 8. Eukaryot Cell. 2009. PMID: 19429778 Free PMC article.
-
Localization of ASH1 mRNA particles in living yeast.Mol Cell. 1998 Oct;2(4):437-45. doi: 10.1016/s1097-2765(00)80143-4. Mol Cell. 1998. PMID: 9809065
-
RNA localization in yeast: moving towards a mechanism.Biol Cell. 2005 Jan;97(1):75-86. doi: 10.1042/BC20040066. Biol Cell. 2005. PMID: 15601259 Review.
-
Local regulation of mRNA translation: new insights from the bud.Trends Cell Biol. 2008 Mar;18(3):105-11. doi: 10.1016/j.tcb.2007.12.004. Epub 2008 Feb 8. Trends Cell Biol. 2008. PMID: 18262421 Review.
Cited by
-
Two- and Three-Dimensional Tracking of MFA2 mRNA Molecules in Mating Yeast.Cells. 2020 Sep 23;9(10):2151. doi: 10.3390/cells9102151. Cells. 2020. PMID: 32977598 Free PMC article.
-
Hitting the mark: Localization of mRNA and biomolecular condensates in health and disease.Wiley Interdiscip Rev RNA. 2023 Nov-Dec;14(6):e1807. doi: 10.1002/wrna.1807. Epub 2023 Jul 2. Wiley Interdiscip Rev RNA. 2023. PMID: 37393916 Free PMC article. Review.
-
Antagonistic gene transcripts regulate adaptation to new growth environments.Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21087-92. doi: 10.1073/pnas.1111408109. Epub 2011 Dec 12. Proc Natl Acad Sci U S A. 2011. PMID: 22160690 Free PMC article.
-
Inducible control of subcellular RNA localization using a synthetic protein-RNA aptamer interaction.PLoS One. 2012;7(10):e46868. doi: 10.1371/journal.pone.0046868. Epub 2012 Oct 8. PLoS One. 2012. PMID: 23056498 Free PMC article.
-
RNA asymmetric distribution and daughter/mother differentiation in yeast.Curr Opin Microbiol. 2003 Dec;6(6):614-20. doi: 10.1016/j.mib.2003.10.005. Curr Opin Microbiol. 2003. PMID: 14662358 Free PMC article. Review.
References
-
- Barral Y, Mermall V, Mooseker M, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5:841–851. - PubMed
-
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–578. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

