The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis
- PMID: 11553739
- PMCID: PMC117967
- DOI: 10.1104/pp.127.1.108
The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis
Abstract
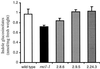
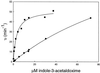
The first committed step in the biosynthesis of indole glucosinolates is the conversion of indole-3-acetaldoxime into an indole-3-S-alkyl-thiohydroximate. The initial step in this conversion is catalyzed by CYP83B1 in Arabidopsis (S. Bak, F.E. Tax, K.A. Feldmann, D.A. Galbraith, R. Feyereisen [2001] Plant Cell 13: 101-111). The knockout mutant of the CYP83B1 gene (rnt1-1) shows a strong auxin excess phenotype and are allelic to sur-2. CYP83A1 is the closest relative to CYP83B1 and shares 63% amino acid sequence identity. Although expression of CYP83A1 under control of its endogenous promoter in the rnt1-1 background does not prevent the auxin excess and indole glucosinolate deficit phenotype caused by the lack of the CYP83B1 gene, ectopic overexpression of CYP83A1 using a 35S promoter rescues the rnt1-1 phenotype. CYP83A1 and CYP83B1 heterologously expressed in yeast (Saccharomyces cerevisiae) cells show marked differences in their substrate specificity. Both enzymes convert indole-3-acetaldoxime to a thiohydroximate adduct in the presence of NADPH and a nucleophilic thiol donor. However, indole-3-acetaldoxime has a 50-fold higher affinity toward CYP83B1 than toward CYP83A1. Both enzymes also metabolize the phenylalanine- and tyrosine-derived aldoximes. Enzyme kinetic comparisons of CYP83A1 and CYP83B1 show that indole-3-acetaldoxime is the physiological substrate for CYP83B1 but not for CYP83A1. Instead, CYP83A1 catalyzes the initial conversion of aldoximes to thiohydroximates in the synthesis of glucosinolates not derived from tryptophan. The two closely related CYP83 subfamily members therefore are not redundant. The presence of putative auxin responsive cis-acting elements in the CYP83B1 promoter but not in the CYP83A1 promoter supports the suggestion that CYP83B1 has evolved to selectively metabolize a tryptophan-derived aldoxime intermediate shared with the pathway of auxin biosynthesis in Arabidopsis.
Figures




References
-
- Andersen AS, Muir R. Auxin activity of glucobrassicin. Plant Physiol. 1966;19:1038–1048.
-
- Andersen MD, Busk PK, Svendsen I, Møller BL. Cytochromes P450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin: cloning, functional expression in Pichia pastoris, and substrate specificity of the isolated recombinant enzymes. J Biol Chem. 2000;275:1966–1977. - PubMed
-
- Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA. Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol. 1998a;36:393–405. - PubMed
-
- Bak S, Nielsen HL, Halkier BA. The presence of CYP79 homologoues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acids to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol Biol. 1998b;38:725–734. - PubMed
-
- Bak S, Olsen CE, Petersen BL, Møller BL, Halkier BA. Metabolic engineering of p-hydroxybenzylglucosinolate in Arabidopsis by expression of the cyanogenic CYP79A1 from Sorghum bicolor. Plant J. 1999;20:663–672. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

