Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes
- PMID: 11553742
- PMCID: PMC117970
- DOI: 10.1104/pp.127.1.142
Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes
Abstract
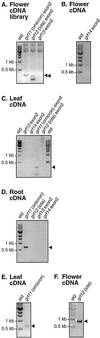
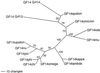
In plants, 14-3-3 proteins are key regulators of primary metabolism and membrane transport. Although the current dogma states that 14-3-3 isoforms are not very specific with regard to target proteins, recent data suggest that the specificity may be high. Therefore, identification and characterization of all 14-3-3 (GF14) isoforms in the model plant Arabidopsis are important. Using the information now available from The Arabidopsis Information Resource, we found three new GF14 genes. The potential expression of these three genes, and of two additional novel GF14 genes (Rosenquist et al., 2000), in leaves, roots, and flowers was examined using reverse transcriptase-polymerase chain reaction and cDNA library polymerase chain reaction screening. Under normal growth conditions, two of these genes were found to be transcribed. These genes were named grf11and grf12, and the corresponding new 14-3-3 isoforms were named GF14omicron and GF14iota, respectively. The gene coding for GF14omicron was expressed in leaves, roots, and flowers, whereas the gene coding for GF14iota was only expressed in flowers. Gene structures and relationships between all members of the GF14 gene family were deduced from data available through The Arabidopsis Information Resource. The data clearly support the theory that two 14-3-3 genes were present when eudicotyledons diverged from monocotyledons. In total, there are 15 14-3-3 genes (grfs 1-15) in Arabidopsis, of which 12 (grfs 1-12) now have been shown to be expressed.
Figures



Similar articles
-
Molecular organization and tissue-specific expression of an Arabidopsis 14-3-3 gene.Plant Cell. 1996 Aug;8(8):1239-48. doi: 10.1105/tpc.8.8.1239. Plant Cell. 1996. PMID: 8776894 Free PMC article.
-
The Arabidopsis 14-3-3 multigene family.Plant Physiol. 1997 Aug;114(4):1421-31. doi: 10.1104/pp.114.4.1421. Plant Physiol. 1997. PMID: 9276953 Free PMC article.
-
Several distinct genes encode nearly identical to 16 kDa proteolipids of the vacuolar H(+)-ATPase from Arabidopsis thaliana.Plant Mol Biol. 1995 Oct;29(2):227-44. doi: 10.1007/BF00043648. Plant Mol Biol. 1995. PMID: 7579175
-
Regulatory 14-3-3 protein-protein interactions in plant cells.Curr Opin Plant Biol. 2000 Oct;3(5):400-5. doi: 10.1016/s1369-5266(00)00103-5. Curr Opin Plant Biol. 2000. PMID: 11019808 Review.
-
The 14-3-3s.Genome Biol. 2002 Jun 27;3(7):REVIEWS3010. doi: 10.1186/gb-2002-3-7-reviews3010. Epub 2002 Jun 27. Genome Biol. 2002. PMID: 12184815 Free PMC article. Review.
Cited by
-
Genome-Wide Identification and Expression Analysis of the 14-3-3 Family Genes in Medicago truncatula.Front Plant Sci. 2016 Mar 22;7:320. doi: 10.3389/fpls.2016.00320. eCollection 2016. Front Plant Sci. 2016. PMID: 27047505 Free PMC article.
-
Plant 14-3-3 proteins as spiders in a web of phosphorylation.Protoplasma. 2013 Apr;250(2):425-40. doi: 10.1007/s00709-012-0437-z. Epub 2012 Aug 29. Protoplasma. 2013. PMID: 22926776 Review.
-
The Glutamate Receptor-Like Protein GLR3.7 Interacts With 14-3-3ω and Participates in Salt Stress Response in Arabidopsis thaliana.Front Plant Sci. 2019 Sep 30;10:1169. doi: 10.3389/fpls.2019.01169. eCollection 2019. Front Plant Sci. 2019. PMID: 31632419 Free PMC article.
-
Identification and Expression Analysis of Wheat TaGF14 Genes.Front Genet. 2018 Jan 30;9:12. doi: 10.3389/fgene.2018.00012. eCollection 2018. Front Genet. 2018. PMID: 29441089 Free PMC article.
-
14-3-3σ and Its Modulators in Cancer.Pharmaceuticals (Basel). 2020 Dec 3;13(12):441. doi: 10.3390/ph13120441. Pharmaceuticals (Basel). 2020. PMID: 33287252 Free PMC article. Review.
References
-
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC. 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 1996;398:26–30. - PubMed
-
- Brandt J, Thordal-Christensen H, Vad K, Gregersen PL, Collinge DB. A pathogen-induced gene of barley encodes a protein showing high similarity to a protein kinase regulator. Plant J. 1992;2:815–820. - PubMed
-
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116.
-
- Finnie C, Borch J, Collinge DB. 14-3-3 proteins: eukaryotic regulatory proteins with many functions. Plant Mol Biol. 1999;40:545–554. - PubMed
-
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

