SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein
- PMID: 11553772
- PMCID: PMC58726
- DOI: 10.1073/pnas.191361798
SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein
Abstract
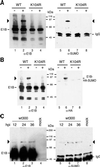
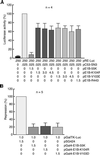
SUMO-1 is a small ubiquitin-related modifier protein that is covalently linked to many cellular and viral protein targets. Modification by SUMO-1 is proposed to play a role in protein targeting and/or stability. We show here that adenovirus type 5 early region 1B 55-kDa (E1B-55kDa) oncoprotein can be covalently modified by SUMO-1 in vivo through a major attachment site comprising a single lysine residue at amino acid position 104. The sequence surrounding this lysine matches the proposed PsiKxE consensus motif required for SUMO-1 conjugation. A single mutation (K104R) that abolishes SUMOylation of E1B-55kDa dramatically reduces the ability of the adenovirus type 5 protein to transform primary baby rat kidney cells in cooperation with E1A and to inhibit p53-mediated transactivation. Overexpression of SUMO-1 in adenovirus type 5 E1A/E1B-55kDa-transformed baby rat kidney cells causes the relocalization of E1B-55kDa from the cytoplasm to the nucleus, where it accumulates with SUMO-1 in dot- or track-like structures. Significantly, when SUMO-1 is ectopically expressed in transformed rat cells no effect on the cytoplasmic localization of the E1B-K104R mutant protein is observed. Our results demonstrate that SUMO-1 modification is required for transformation by adenovirus type 5 E1B-55kDa and provide further evidence for the idea that this posttranslational modification plays a role in protein targeting to specific subcellular sites.
Figures




Similar articles
-
Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1.EMBO J. 1999 Nov 15;18(22):6462-71. doi: 10.1093/emboj/18.22.6462. EMBO J. 1999. PMID: 10562558 Free PMC article.
-
Blockage of CRM1-dependent nuclear export of the adenovirus type 5 early region 1B 55-kDa protein augments oncogenic transformation of primary rat cells.Oncogene. 2005 Jan 6;24(1):55-64. doi: 10.1038/sj.onc.1208170. Oncogene. 2005. PMID: 15480414
-
Adenovirus type 5 early region 1B 55-kDa oncoprotein can promote cell transformation by a mechanism independent from blocking p53-activated transcription.Oncogene. 2008 Jun 12;27(26):3673-84. doi: 10.1038/sj.onc.1211039. Epub 2008 Jan 21. Oncogene. 2008. PMID: 18212738
-
SUMO conjugation in plants.Planta. 2004 Nov;220(1):1-8. doi: 10.1007/s00425-004-1370-y. Epub 2004 Sep 23. Planta. 2004. PMID: 15449058 Review.
-
Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity.Curr Opin Genet Dev. 2003 Apr;13(2):108-13. doi: 10.1016/s0959-437x(03)00021-2. Curr Opin Genet Dev. 2003. PMID: 12672486 Review.
Cited by
-
The human adenovirus type 5 E1B 55 kDa protein obstructs inhibition of viral replication by type I interferon in normal human cells.PLoS Pathog. 2012;8(8):e1002853. doi: 10.1371/journal.ppat.1002853. Epub 2012 Aug 9. PLoS Pathog. 2012. PMID: 22912576 Free PMC article.
-
Efficient Transformation of Primary Human Mesenchymal Stromal Cells by Adenovirus Early Region 1 Oncogenes.J Virol. 2016 Dec 16;91(1):e01782-16. doi: 10.1128/JVI.01782-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795433 Free PMC article.
-
Timely synthesis of the adenovirus type 5 E1B 55-kilodalton protein is required for efficient genome replication in normal human cells.J Virol. 2012 Mar;86(6):3064-72. doi: 10.1128/JVI.06764-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278242 Free PMC article.
-
SCE1, the SUMO-conjugating enzyme in plants that interacts with NIb, the RNA-dependent RNA polymerase of Turnip mosaic virus, is required for viral infection.J Virol. 2013 Apr;87(8):4704-15. doi: 10.1128/JVI.02828-12. Epub 2013 Jan 30. J Virol. 2013. PMID: 23365455 Free PMC article.
-
Adenovirus type 5 early region 1B 156R protein promotes cell transformation independently of repression of p53-stimulated transcription.J Virol. 2007 Jan;81(1):95-105. doi: 10.1128/JVI.01608-06. Epub 2006 Oct 18. J Virol. 2007. PMID: 17050591 Free PMC article.
References
-
- Shenk T. In: Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. New York: Lippincott-Raven; 1996. pp. 2111–2148.
-
- Barker D D, Berk A J. Virology. 1987;156:107–121. - PubMed
-
- Sarnow P, Ho Y S, Williams J, Levine A J. Cell. 1982;28:387–394. - PubMed
-
- Yew P R, Berk A J. Nature (London) 1992;357:82–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

