Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: studies of fusion proteins
- PMID: 11553789
- PMCID: PMC58691
- DOI: 10.1073/pnas.201387098
Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: studies of fusion proteins
Abstract
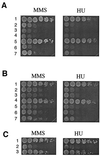
The Sgs1 protein of the budding yeast Saccharomyces cerevisiae is a member of the RecQ DNA helicase family that includes the human Bloom, Werner, and Rothmund-Thompson syndrome proteins. The N-terminal region outside the central DNA helicase core of Sgs1, particularly the part containing the first 100 amino acid residues of the 1,447-residue protein, is known to be functionally important and has been implicated in Sgs1-DNA topoisomerase III (Top3) interaction. We show in this work that the functionality of a truncated Sgs1 lacking its N-terminal 106 residues can be restored by replacing the truncated region with Top3. Fusion of Top3 to a mutant Sgs1 with a Val-29 to Glu substitution, which interferes with Sgs1-Top3 interaction, similarly restores the functionality of the mutant Sgs1(V29E) protein. The Top3-Sgs1(Delta1-106) and Top3-Sgs1(V29E) fusion proteins behave like wild-type Sgs1 in complementing several aspects of the sgs1 phenotype, including the hypersensitivity of sgs1 cells to methyl methanesulfonate and hydroxyurea. Complementation by the fusion proteins required both the topoisomerase activity of Top3 and the helicase activity of the Sgs1 polypeptide. These results suggest that the sole function of the N-terminal 106 amino acid residues of Sgs1 is for Top3 binding, and that the coordinated actions of Sgs1 and Top3 are important in cellular processes such as the processing of DNA after exposure of cells to DNA-damaging agents.
Figures



References
-
- Enomoto T. J Biochem (Tokyo) 2001;129:501–507. - PubMed
-
- Frei C, Gasser S M. J Cell Sci. 2000;113:2641–2646. - PubMed
-
- Karow J K, Wu L, Hickson I D. Curr Opin Genet Dev. 2000;10:32–38. - PubMed
-
- Shen J C, Loeb L A. Trends Genet. 2000;16:213–220. - PubMed
-
- Ellis N A, Groden J, Ye T-Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

