DNA microarrays in medical practice
- PMID: 11557712
- PMCID: PMC1121185
- DOI: 10.1136/bmj.323.7313.611
DNA microarrays in medical practice
Figures



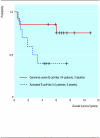
Similar articles
-
[A prospect of genetic tests].Rinsho Byori. 2003 Jun;Suppl 126:139-44. Rinsho Byori. 2003. PMID: 12905954 Review. Japanese. No abstract available.
-
[DNA chips--a new diagnostic revolution?].Duodecim. 1998;114(9):829, 831. Duodecim. 1998. PMID: 11524802 Finnish. No abstract available.
-
Types of array findings detectable in cytogenetic diagnosis: a proposal for a generic classification.Eur J Hum Genet. 2014 Jul;22(7):856-8. doi: 10.1038/ejhg.2013.254. Epub 2013 Nov 6. Eur J Hum Genet. 2014. PMID: 24193341 Free PMC article. Review. No abstract available.
-
Oligonucleotide microarrays for clinical diagnosis of copy number variation and zygosity status.Curr Protoc Hum Genet. 2012 Jul;Chapter 8:Unit8.12. doi: 10.1002/0471142905.hg0812s74. Curr Protoc Hum Genet. 2012. PMID: 22786613
-
[Gene diagnosis and gene therapy].Nihon Naika Gakkai Zasshi. 1993 Apr 10;82(4):598-604. Nihon Naika Gakkai Zasshi. 1993. PMID: 8340672 Review. Japanese. No abstract available.
Cited by
-
Genetics 101: detecting mutations in human genes.CMAJ. 2002 Aug 6;167(3):275-9. CMAJ. 2002. PMID: 12186176 Free PMC article. Review. No abstract available.
-
Application of microarray technology in pulmonary diseases.Respir Res. 2004 Dec 7;5(1):26. doi: 10.1186/1465-9921-5-26. Respir Res. 2004. PMID: 15585067 Free PMC article. Review.
-
Molecular genetics of Psoriasis (Principles, technology, gene location, genetic polymorphism and gene expression).Int J Health Sci (Qassim). 2010 Nov;4(2):103-27. Int J Health Sci (Qassim). 2010. PMID: 21475550 Free PMC article.
-
Molecular imaging in oncology.Cancer Imaging. 2004 Oct 21;4(2):162-73. doi: 10.1102/1470-7330.2004.0060. Cancer Imaging. 2004. PMID: 18250026 Free PMC article.
-
Neurovirological methods and their applications.J Neurol Neurosurg Psychiatry. 2003 Aug;74(8):1016-22. doi: 10.1136/jnnp.74.8.1016. J Neurol Neurosurg Psychiatry. 2003. PMID: 12876227 Free PMC article. Review.
References
-
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
-
- Collins FS. Shattuck lecture—medical and societal consequences of the human genome project. N Engl J Med. 1999;341:28–37. - PubMed
-
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nature Genet. 1999;21(suppl):33–37. - PubMed
-
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
