Structure and function of type II restriction endonucleases
- PMID: 11557805
- PMCID: PMC55916
- DOI: 10.1093/nar/29.18.3705
Structure and function of type II restriction endonucleases
Abstract
More than 3000 type II restriction endonucleases have been discovered. They recognize short, usually palindromic, sequences of 4-8 bp and, in the presence of Mg(2+), cleave the DNA within or in close proximity to the recognition sequence. The orthodox type II enzymes are homodimers which recognize palindromic sites. Depending on particular features subtypes are classified. All structures of restriction enzymes show a common structural core comprising four beta-strands and one alpha-helix. Furthermore, two families of enzymes can be distinguished which are structurally very similar (EcoRI-like enzymes and EcoRV-like enzymes). Like other DNA binding proteins, restriction enzymes are capable of non-specific DNA binding, which is the prerequisite for efficient target site location by facilitated diffusion. Non-specific binding usually does not involve interactions with the bases but only with the DNA backbone. In contrast, specific binding is characterized by an intimate interplay between direct (interaction with the bases) and indirect (interaction with the backbone) readout. Typically approximately 15-20 hydrogen bonds are formed between a dimeric restriction enzyme and the bases of the recognition sequence, in addition to numerous van der Waals contacts to the bases and hydrogen bonds to the backbone, which may also be water mediated. The recognition process triggers large conformational changes of the enzyme and the DNA, which lead to the activation of the catalytic centers. In many restriction enzymes the catalytic centers, one in each subunit, are represented by the PD. D/EXK motif, in which the two carboxylates are responsible for Mg(2+) binding, the essential cofactor for the great majority of enzymes. The precise mechanism of cleavage has not yet been established for any enzyme, the main uncertainty concerns the number of Mg(2+) ions directly involved in cleavage. Cleavage in the two strands usually occurs in a concerted fashion and leads to inversion of configuration at the phosphorus. The products of the reaction are DNA fragments with a 3'-OH and a 5'-phosphate.
Figures
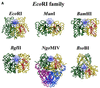
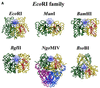




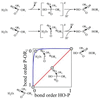

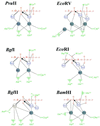
References
-
- Roberts R.J. and Halford,S.E. (1993) Type II restriction endonucleases. In Linn,S.M., Lloyd,R.S. and Roberts,R.J. (eds), Nucleases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY, pp. 35–88.
-
- Raleigh E.A. and Brooks,J.E. (1998) In De Bruijn,F.J., Lupski,J.R. and Weinstock,G.M. (eds), Bacterial Genomes. Chapman & Hall, New York, pp. 78–92.
-
- Arber W. (1979) Promotion and limitation of genetic exchange. Science, 205, 361–365. - PubMed
-
- Heitman J. (1993) On the origins, structures and functions of restriction-modification enzymes. In Setlow,J.K. (ed.), Genetic Engineering. Plenum Press, New York, Vol. 15, pp. 57–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

