Molecular cloning of a cDNA encoding mouse DNA helicase B, which has homology to Escherichia coli RecD protein, and identification of a mutation in the DNA helicase B from tsFT848 temperature-sensitive DNA replication mutant cells
- PMID: 11557815
- PMCID: PMC55905
- DOI: 10.1093/nar/29.18.3835
Molecular cloning of a cDNA encoding mouse DNA helicase B, which has homology to Escherichia coli RecD protein, and identification of a mutation in the DNA helicase B from tsFT848 temperature-sensitive DNA replication mutant cells
Abstract
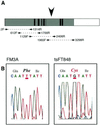
DNA helicase B is a major DNA helicase in mouse FM3A cells. A temperature-sensitive mutant defective in DNA replication, tsFT848, isolated from FM3A cells, has a heat-labile DNA helicase B. In this study, we purified DNA helicase B from mouse FM3A cells and determined partial amino acid sequences of the purified protein. By using a DNA probe synthesized according to one of the partial amino acid sequences, a cDNA was isolated, which encoded a 121.5 kDa protein containing seven conserved motifs for DNA/RNA helicase superfamily members. A database search revealed similarity between DNA helicase B and the alpha subunit of exodeoxyribonuclease V of a number of prokaryotes including Escherichia coli RecD protein, but no homologous protein was found in yeast. The cDNA encoding DNA helicase B from tsFT848 was sequenced and a mutation was found between DNA/RNA helicase motifs IV and V.
Figures

References
-
- Matson S.W. and Kaiser-Rogers,K.A. (1990) DNA helicases. Annu. Rev. Biochem., 59, 289–329. - PubMed
-
- Weeda G., van Ham,R.C., Vermeulen,W., Bootsma,D., van der Eb,A.J. and Hoeijmakers,J.H. (1990) A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne’s syndrome. Cell, 62, 777–791. - PubMed
-
- Schaeffer L., Roy,R., Humbert,S., Moncollin,V., Vermeulen,W., Hoeijmakers,J.H., Chambon,P. and Egly,J.M. (1993) DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science, 260, 58–63. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

