Differentiation-dependent chromatin rearrangement coincides with activation of human papillomavirus type 31 late gene expression
- PMID: 11559836
- PMCID: PMC114575
- DOI: 10.1128/JVI.75.20.10005-10013.2001
Differentiation-dependent chromatin rearrangement coincides with activation of human papillomavirus type 31 late gene expression
Abstract
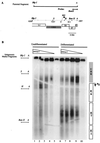
The life cycle of human papillomaviruses (HPVs) is tightly linked to the differentiation status of the host cell. While early genes are expressed during the initial stages of viral infection, late gene expression occurs in the suprabasal layers of the cervical epithelium. Late genes encode E1-E4, a cytosolic protein, and capsid proteins L1 and L2. We have mapped over 30 initiation sites for late transcripts and show that the transcripts initiate in a 200-nucleotide region within the E7 open reading frame. The mechanisms regulating the activation of late gene expression, however, are not yet understood. DNase I hypersensitivity analysis of HPV-31 chromatin in cell lines that maintain viral genomes extrachromosomally indicates that a major shift in nuclease digestion occurs upon differentiation. In undifferentiated cells, hypersensitive regions exist in the upstream regulatory region proximal to the E6 open reading frame. Upon differentiation, a region between nucleotides 659 and 811 in the E7 open reading frame becomes accessible to DNase I. These results indicate that the late transcript initiation region becomes accessible to transcription factor binding upon differentiation. Several complexes mediate chromatin rearrangement, and we tested whether histone acetylation was sufficient for late transcript activation. Treatment with the histone deacetylase inhibitor trichostatin A was found to be insufficient to activate late gene expression in undifferentiated cells. However, it did activate expression of early transcripts. These results suggest that chromatin remodeling around the late promoter occurs upon epithelial differentiation and that mechanisms in addition to histone deacetylation contribute to activation of late gene expression.
Figures

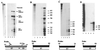


References
-
- Ait-Si-Ali S, Polesskaya A, Filleur S, Ferreira R, Duquet A, Robin P, Vervish A, Trouche D, Cabon F, Harel-Bellan A. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene. 2000;19:2430–2437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

