Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo
- PMID: 11562474
- PMCID: PMC58734
- DOI: 10.1073/pnas.201284398
Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo
Abstract
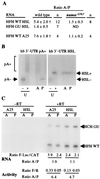
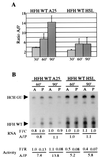
Development of the Drosophila abdomen requires repression of maternal hunchback (hb) mRNA translation in the posterior of the embryo. This regulation involves at least four components: nanos response elements within the hb 3' untranslated region and the activities of Pumilio (PUM), Nanos (NOS), and Brain tumor. To study this regulation, we have developed an RNA injection assay that faithfully recapitulates the regulation of the endogenous hb message. Previous studies have suggested that NOS and PUM can regulate translation by directing poly(A) removal. We have found that RNAs that lack a poly(A) tail and cannot be polyadenylated and RNAs that contain translational activating sequences in place of the poly(A) tail are still repressed in the posterior. These data demonstrate that the poly(A) tail is not required for regulation and suggest that NOS and PUM can regulate hb translation by two mechanisms: removal of the poly(A) tail and a poly(A)-independent pathway that directly affects translation.
Figures


References
-
- Macdonald P M, Smibert C A. Curr Opin Genet Dev. 1996;6:403–407. - PubMed
-
- Sheets M D, Fox C A, Hunt T, Vande Woude G, Wickens M. Genes Dev. 1994;8:926–938. - PubMed
-
- Salles F J, Lieberfarb M E, Wreden C, Gergen J P, Strickland S. Science. 1994;266:1996–1999. - PubMed
-
- Lieberfarb M E, Chu T, Wreden C, Theurkauf W, Gergen J P, Strickland S. Development (Cambridge, UK) 1996;122:579–588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

