Use of chromatin immunoprecipitation to clone novel E2F target promoters
- PMID: 11564866
- PMCID: PMC99859
- DOI: 10.1128/MCB.21.20.6820-6832.2001
Use of chromatin immunoprecipitation to clone novel E2F target promoters
Abstract
We have taken a new approach to the identification of E2F-regulated promoters. After modification of a chromatin immunoprecipitation assay, we cloned nine chromatin fragments which represent both strong and weak in vivo E2F binding sites. Further characterization of three of the cloned fragments revealed that they are bound in vivo not only by E2Fs but also by members of the retinoblastoma tumor suppressor protein family and by RNA polymerase II, suggesting that these fragments represent promoters regulated by E2F transcription complexes. In fact, database analysis indicates that all three fragments correspond to genomic DNA located just upstream of start sites for previously identified mRNAs. One clone, ChET 4, corresponds to the promoter region for beclin 1, a candidate tumor suppressor protein. We demonstrate that another of the clones, ChET 8, is strongly bound by E2F family members in vivo but does not contain a consensus E2F binding site. However, this fragment functions as a promoter whose activity can be repressed by E2F1. Finally, we demonstrate that the ChET 9 promoter contains a consensus E2F binding site, can be activated by E2F1, and drives expression of an mRNA that is upregulated in colon and liver tumors. Interestingly, the characterized ChET promoters do not display regulation patterns typical of known E2F target genes in a U937 cell differentiation system. In summary, we have provided evidence that chromatin immunoprecipitation can be used to identify E2F-regulated promoters which contain both consensus and nonconsensus binding sites and have shown that not all E2F-regulated promoters show identical expression profiles.
Figures
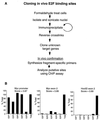
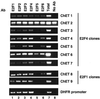



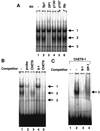


References
-
- Aita V M, Liang X H, Murty V V, Pincus D L, Yu W, Cayanis E, Kalachikov S, Gilliam T C, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. - PubMed
-
- Albert T, Wells J, Funk J-O, Pullner A, Raschke E-E, Stelzer G, Meisterernst M, Farnham P J, Eick D. The chromatin structure of the dual c-Myc promoter P1/P2 is regulated by separate elements. J Biol Chem. 2001;276:20482–20490. - PubMed
-
- Bussemaker H J, Li H, Siggia E D. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
