Differential interactions of specific nuclear factor I isoforms with the glucocorticoid receptor and STAT5 in the cooperative regulation of WAP gene transcription
- PMID: 11564870
- PMCID: PMC99863
- DOI: 10.1128/MCB.21.20.6859-6869.2001
Differential interactions of specific nuclear factor I isoforms with the glucocorticoid receptor and STAT5 in the cooperative regulation of WAP gene transcription
Abstract
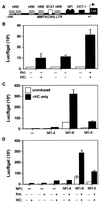
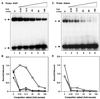
The distal region (-830 to -720 bp) of the rat whey acidic protein (WAP) gene contains a composite response element (CoRE), which has been demonstrated previously to confer mammary gland-specific and hormonally regulated WAP gene expression. Point mutations in the binding sites for specific transcription factors present within this CoRE have demonstrated the importance of both nuclear factor I (NFI) and STAT5 as well as cooperative interactions with the glucocorticoid receptor (GR) in the regulation of WAP gene expression in the mammary gland of transgenic mice. This study reports the characterization of NFI gene expression during mammary gland development and the identification and cloning of specific NFI isoforms (NFI-A4, NFI-B2, and NFI-X1) from the mouse mammary gland during lactation. Some but not all of these NFI isoforms synergistically activate WAP gene transcription in cooperation with GR and STAT5, as determined using transient cotransfection assays in JEG-3 cells. On both the WAP CoRE and the mouse mammary tumor virus long terminal repeat promoter, the NFI-B isoform preferentially activated gene transcription in cooperation with STAT5A and GR. In contrast, the NFI-A isoform suppressed GR and STAT cooperativity at the WAP CoRE. Finally, unlike their interaction with the NFI consensus binding site in the adenovirus promoter, the DNA-binding specificities of the three NFI isoforms to the palindromic NFI site in the WAP CoRE were not identical, which may partially explain the failure of the NFI-A isoform to cooperate with GR and STAT5A.
Figures




Similar articles
-
The C-terminal domain of the nuclear factor I-B2 isoform is glycosylated and transactivates the WAP gene in the JEG-3 cells.Biochem Biophys Res Commun. 2007 Jul 6;358(3):770-6. doi: 10.1016/j.bbrc.2007.04.185. Epub 2007 May 7. Biochem Biophys Res Commun. 2007. PMID: 17511965 Free PMC article.
-
Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in transgenic mice.Mol Cell Biol. 1995 Apr;15(4):2063-70. doi: 10.1128/MCB.15.4.2063. Mol Cell Biol. 1995. PMID: 7891701 Free PMC article.
-
Composite response elements mediate hormonal and developmental regulation of milk protein gene expression.Biochem Soc Symp. 1998;63:101-13. Biochem Soc Symp. 1998. PMID: 9513715 Review.
-
Constitutive repression and nuclear factor I-dependent hormone activation of the mouse mammary tumor virus promoter in Saccharomyces cerevisiae.Mol Cell Biol. 1995 Dec;15(12):6987-98. doi: 10.1128/MCB.15.12.6987. Mol Cell Biol. 1995. PMID: 8524266 Free PMC article.
-
Synergistic and antagonistic interactions of transcription factors in the regulation of milk protein gene expression. Mechanisms of cross-talk between signalling pathways.Adv Exp Med Biol. 2000;480:139-46. doi: 10.1007/0-306-46832-8_17. Adv Exp Med Biol. 2000. PMID: 10959420 Review.
Cited by
-
Redundant and non-redundant cytokine-activated enhancers control Csn1s2b expression in the lactating mouse mammary gland.Nat Commun. 2021 Apr 14;12(1):2239. doi: 10.1038/s41467-021-22500-w. Nat Commun. 2021. PMID: 33854063 Free PMC article.
-
Polycystin-1 regulates skeletogenesis through stimulation of the osteoblast-specific transcription factor RUNX2-II.J Biol Chem. 2008 May 2;283(18):12624-34. doi: 10.1074/jbc.M710407200. Epub 2008 Mar 5. J Biol Chem. 2008. PMID: 18321855 Free PMC article.
-
The C-terminal domain of the nuclear factor I-B2 isoform is glycosylated and transactivates the WAP gene in the JEG-3 cells.Biochem Biophys Res Commun. 2007 Jul 6;358(3):770-6. doi: 10.1016/j.bbrc.2007.04.185. Epub 2007 May 7. Biochem Biophys Res Commun. 2007. PMID: 17511965 Free PMC article.
-
Transcriptional regulation of the cyclin-dependent kinase inhibitor 1A (p21) gene by NFI in proliferating human cells.Nucleic Acids Res. 2006;34(22):6472-87. doi: 10.1093/nar/gkl861. Epub 2006 Nov 27. Nucleic Acids Res. 2006. PMID: 17130157 Free PMC article.
-
A 470 bp WAP-promoter fragment confers lactation independent, progesterone regulated mammary-specific gene expression in transgenic mice.Transgenic Res. 2005 Apr;14(2):145-58. doi: 10.1007/s11248-004-7434-8. Transgenic Res. 2005. PMID: 16022386
References
-
- Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. . (Erratum, 256:161.) - PubMed
-
- Bachurski C J, Kelly S E, Glasser S W, Currier T A. Nuclear factor I family members regulate the transcription of surfactant protein-C. J Biol Chem. 1997;272:32759–32766. - PubMed
-
- Bandyopadhyay S, Starke D W, Mieyal J J, Gronostajski R M. Thioltransferase (glutaredoxin) reactivates the DNA-binding activity of oxidation-inactivated nuclear factor I. J Biol Chem. 1998;273:392–397. - PubMed
-
- Blum J L, Zeigler M E, Wicha M S. Regulation of rat mammary gene expression by extracellular matrix components. Exp Cell Res. 1987;173:322–340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
