Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae
- PMID: 11564888
- PMCID: PMC99881
- DOI: 10.1128/MCB.21.20.7054-7064.2001
Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae
Abstract
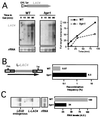
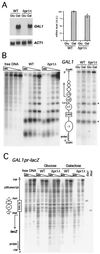
Hpr1 forms, together with Tho2, Mft1, and Thp2, the THO complex, which controls transcription elongation and genome stability in Saccharomyces cerevisiae. Mutations in genes encoding the THO complex confer strong transcription-impairment and hyperrecombination phenotypes in the bacterial lacZ gene. In this work we demonstrate that Hpr1 is a factor required for transcription of long as well as G+C-rich DNA sequences. Using different lacZ segments fused to the GAL1 promoter, we show that the negative effect of lacZ sequences on transcription depends on their distance from the promoter. In parallel, we show that transcription of either a long LYS2 fragment or the S. cerevisiae YAT1 G+C-rich open reading frame fused to the GAL1 promoter is severely impaired in hpr1 mutants, whereas transcription of LAC4, the Kluyveromyces lactis ortholog of lacZ but with a lower G+C content, is only slightly affected. The hyperrecombination behavior of the DNA sequences studied is consistent with the transcriptional defects observed in hpr1 cells. These results indicate that both length and G+C content are important elements influencing transcription in vivo. We discuss their relevance for the understanding of the functional role of Hpr1 and, by extension, the THO complex.
Figures
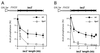






References
-
- Adam A C, Prieto J A, Rubio-Texeira M, Polaina J. Construction of a lactose-assimilating strain of baker's yeast. Yeast. 1999;15:1299–1305. - PubMed
-
- Aguilera A, Chávez S S, Malagón F. Mitotic recombination in yeast: elements controlling its incidence. Yeast. 2000;16:731–754. - PubMed
-
- Aso T, Lane W S, Conaway J W, Conaway R C. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
