The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression
- PMID: 11564889
- PMCID: PMC99882
- DOI: 10.1128/MCB.21.20.7065-7077.2001
The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression
Abstract
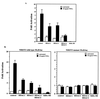
Regulation of NF-kappaB transactivation function is controlled at several levels, including interactions with coactivator proteins. Here we show that the transactivation function of NF-kappaB is also regulated through interaction of the p65 (RelA) subunit with histone deacetylase (HDAC) corepressor proteins. Our results show that inhibition of HDAC activity with trichostatin A (TSA) results in an increase in both basal and induced expression of an integrated NF-kappaB-dependent reporter gene. Chromatin immunoprecipitation (ChIP) assays show that TSA treatment causes hyperacetylation of the wild-type integrated NF-kappaB-dependent reporter but not of a mutant version in which the NF-kappaB binding sites were mutated. Expression of HDAC1 and HDAC2 repressed tumor necrosis factor (TNF)-induced NF-kappaB-dependent gene expression. Consistent with this, we show that HDAC1 and HDAC2 target NF-kappaB through a direct association of HDAC1 with the Rel homology domain of p65. HDAC2 does not interact with NF-kappaB directly but can regulate NF-kappaB activity through its association with HDAC1. Finally, we show that inhibition of HDAC activity with TSA causes an increase in both basal and TNF-induced expression of the NF-kappaB-regulated interleukin-8 (IL-8) gene. Similar to the wild-type integrated NF-kappaB-dependent reporter, ChIP assays showed that TSA treatment resulted in hyperacetylation of the IL-8 promoter. These data indicate that the transactivation function of NF-kappaB is regulated in part through its association with HDAC corepressor proteins. Moreover, it suggests that the association of NF-kappaB with the HDAC1 and HDAC2 corepressor proteins functions to repress expression of NF-kappaB-regulated genes as well as to control the induced level of expression of these genes.
Figures



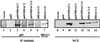



References
-
- Ashburner B P, Shackelford R E, Baldwin A S, Jr, Paules R S. Lack of involvement of ataxia telangiectasia mutated (ATM) in regulation of nuclear factor-kappaB (NF-kappaB) in human diploid fibroblasts. Cancer Res. 1999;59:5456–5460. - PubMed
-
- Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. - PubMed
-
- Baldwin A S., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. - PubMed
-
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Carlotti F, Dower S K, Qwarnstrom E E. Dynamic shuttling of nuclear factor kappa B between the nucleus and cytoplasm as a consequence of inhibitor dissociation. J Biol Chem. 2000;275:41028–41034. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
