Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site
- PMID: 11566868
- PMCID: PMC125623
- DOI: 10.1093/emboj/20.18.5033
Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site
Abstract
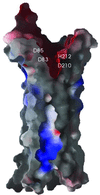
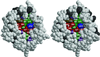
OmpT from Escherichia coli belongs to a family of highly homologous outer membrane proteases, known as omptins, which are implicated in the virulence of several pathogenic Gram-negative bacteria. Here we present the crystal structure of OmpT, which shows a 10-stranded antiparallel beta-barrel that protrudes far from the lipid bilayer into the extracellular space. We identified a putative binding site for lipopolysaccharide, a molecule that is essential for OmpT activity. The proteolytic site is located in a groove at the extracellular top of the vase-shaped beta-barrel. Based on the constellation of active site residues, we propose a novel proteolytic mechanism, involving a His-Asp dyad and an Asp-Asp couple that activate a putative nucleophilic water molecule. The active site is fully conserved within the omptin family. Therefore, the structure described here provides a sound basis for the design of drugs against omptin-mediated bacterial pathogenesis. Coordinates are in the Protein Data Bank (accession No. 1I78)
Figures
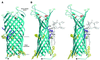




References
-
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Buchanan S.K., Smith,B.S., Venkatramani,L., Xia,D., Esser,L., Palnitkar,M., Chakraborty,R., van der Helm,D. and Deisenhofer,J. (1999) Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nature Struct. Biol., 6, 56–63. - PubMed
-
- Budisa N., Steipe,B., Demange,P., Eckerskorn,C., Kellermann,J. and Huber,R. (1995) High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur. J. Biochem., 230, 788–796. - PubMed
-
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. - PubMed
-
- Cowan S.W., Schirmer,T., Rummel,G., Steiert,M., Ghosh,R., Pauptit, R.A., Jansonius,J.N. and Rosenbusch,J.P. (1992) Crystal structures explain functional properties of two E.coli porins. Nature, 358, 727–733. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

