Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter
- PMID: 11566870
- PMCID: PMC125274
- DOI: 10.1093/emboj/20.18.5049
Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter
Abstract
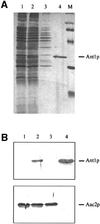
The requirement for small molecule transport systems across the peroxisomal membrane has previously been postulated, but not directly proven. Here we report the identification and functional reconstitution of Ant1p (Ypr128cp), a peroxisomal transporter in the yeast Saccharomyces cerevisiae, which has the characteristic sequence features of the mitochondrial carrier family. Ant1p was found to be an integral protein of the peroxisomal membrane and expression of ANT1 was oleic acid inducible. Targeting of Ant1p to peroxisomes was dependent on Pex3p and Pex19p, two peroxins specifically required for peroxisomal membrane protein insertion. Ant1p was essential for growth on medium-chain fatty acids as the sole carbon source. Upon reconstitution of the overexpressed and purified protein into liposomes, specific transport of adenine nucleotides could be demonstrated. Remarkably, both the substrate and inhibitor specificity differed from those of the mitochondrial ADP/ATP transporter. The physiological role of Ant1p in S.cerevisiae is probably to transport cytoplasmic ATP into the peroxisomal lumen in exchange for AMP generated in the activation of fatty acids.
Figures


References
-
- Blobel F. and Erdmann,R. (1996) Identification of a yeast peroxisomal member of the family of AMP-binding proteins. Eur. J. Biochem., 240, 468–476. - PubMed
-
- Dansen T.B., Wirtz,K.W., Wanders,R.J. and Pap,E.H. (2000) Peroxi somes in human fibroblasts have a basic pH. Nature Cell Biol., 2, 51–53. - PubMed
-
- Daum G., Gasser,S.M. and Schatz,G. (1982) Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J. Biol. Chem., 257, 13075–13080. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

