Net-targeted mutant mice develop a vascular phenotype and up-regulate egr-1
- PMID: 11566878
- PMCID: PMC125619
- DOI: 10.1093/emboj/20.18.5139
Net-targeted mutant mice develop a vascular phenotype and up-regulate egr-1
Abstract
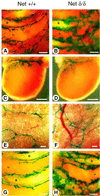
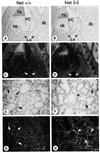
The ternary complex factors (TCFs) Net, Elk-1 and Sap-1 regulate immediate early genes through serum response elements (SREs) in vitro, but, surprisingly, their in vivo roles are unknown. Net is a repressor that is expressed in sites of vasculogenesis during mouse development. We have made gene-targeted mice that express a hypomorphic mutant of Net, Net delta, which lacks the Ets DNA-binding domain. Strikingly, homozygous mutant mice develop a vascular defect and up-regulate an immediate early gene implicated in vascular disease, egr-1. They die after birth due to respiratory failure, resulting from the accumulation of chyle in the thoracic cage (chylothorax). The mice have dilated lymphatic vessels (lymphangiectasis) as early as E16.5. Interestingly, they express more egr-1 in heart and pulmonary arteries at E18.5. Net negatively regulates the egr-1 promoter and binds specifically to SRE-5. Egr-1 has been associated with pathologies involving vascular stenosis (e.g. atherosclerosis), and here egr-1 dysfunction could possibly be associated with obstructions that ultimately affect the lymphatics. These results show that Net is involved in vascular biology and egr-1 regulation in vivo.
Figures










References
-
- al-Moslih M.I. and Dubes,G.R. (1973) The kinetics of DEAE–dextran-induced cell sensitization to transfection. J. Gen. Virol., 18, 189–193. - PubMed
-
- Ayadi A., Suelves,M., Dolle,P. and Wasylyk,B. (2001) Net, an Ets ternary complex transcription factor, is expressed in sites of vasculogenesis, angiogenesis and chondrogenesis during mouse development. Mech. Dev., 102, 205–208. - PubMed
-
- Beddington R.S., Morgernstern,J., Land,H. and Hogan,A. (1989) An in situ transgenic enzyme marker for the midgestation mouse embryo and the visualization of inner cell mass clones during early organogenesis. Development, 106, 37–46. - PubMed
-
- Clarkson R.W., Shang,C.A., Levitt,L.K., Howard,T. and Waters,M.J. (1999) Ternary complex factors Elk-1 and Sap-1a mediate growth hormone-induced transcription of egr-1 (early growth response factor-1) in 3T3-F442A preadipocytes. Mol. Endocrinol., 13, 619–631. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

