Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions
- PMID: 11566884
- PMCID: PMC125637
- DOI: 10.1093/emboj/20.18.5207
Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions
Abstract
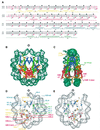
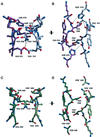
Chromatin is composed of nucleosomes, the universally repeating protein-DNA complex in eukaryotic cells. The crystal structure of the nucleosome core particle from Saccharomyces cerevisiae reveals that the structure and function of this fundamental complex is conserved between single-cell organisms and metazoans. Our results show that yeast nucleosomes are likely to be subtly destabilized as compared with nucleosomes from higher eukaryotes, consistent with the idea that much of the yeast genome remains constitutively open during much of its life cycle. Importantly, minor sequence variations lead to dramatic changes in the way in which nucleosomes pack against each other within the crystal lattice. This has important implications for our understanding of the formation of higher order chromatin structure and its modulation by post-translational modifications. Finally, the yeast nucleosome core particle provides a structural context by which to interpret genetic data obtained from yeast. Coordinates have been deposited with the Protein Data Bank under accession number 1ID3.
Figures


References
-
- Aalfs J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. - PubMed
-
- Baer B.W. and Rhodes,D. (1983) Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature, 301, 482–488. - PubMed
-
- Brünger A.T., Adams,P.D. and Rice,L.M. (1997) New applications of simulated annealing in X-ray crystallography and solution NMR. Structure, 5, 325–336. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

