Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli
- PMID: 11567002
- PMCID: PMC99681
- DOI: 10.1128/JB.183.20.6017-6027.2001
Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli
Erratum in
- J Bacteriol 2002 Feb;184(3):871
Abstract
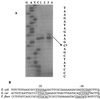
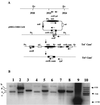
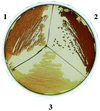
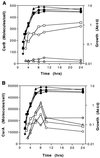
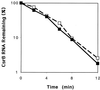
The global regulator CsrA (carbon storage regulator) of Escherichia coli is a small RNA binding protein that represses various metabolic pathways and processes that are induced in the stationary phase of growth, while it activates certain exponential phase functions. Both repression and activation by CsrA involve posttranscriptional mechanisms, in which CsrA binding to mRNA leads to decreased or increased transcript stability, respectively. CsrA also binds to a small untranslated RNA, CsrB, forming a ribonucleoprotein complex, which antagonizes CsrA activity. We have further examined the regulatory interactions of CsrA and CsrB RNA. The 5' end of the CsrB transcript was mapped, and a csrB::cam null mutant was constructed. CsrA protein and CsrB RNA levels were estimated throughout the growth curves of wild-type and isogenic csrA, csrB, rpoS, or csrA rpoS mutant strains. CsrA levels exhibited modest or negligible effects of these mutations. The intracellular concentration of CsrA exceeded the total CsrA-binding capacity of intracellular CsrB RNA. In contrast, CsrB levels were drastically decreased (~10-fold) in the csrA mutants. CsrB transcript stability was unaffected by csrA. The expression of a csrB-lacZ transcriptional fusion containing the region from -242 to +4 bp of the csrB gene was decreased ~20-fold by a csrA::kanR mutation in vivo but was unaffected by CsrA protein in vitro. These results reveal a significant, though most likely indirect, role for CsrA in regulating csrB transcription. Furthermore, our findings suggest that CsrA mediates an intriguing form of autoregulation, whereby its activity, but not its levels, is modulated through effects on an RNA antagonist, CsrB.
Figures


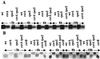


Similar articles
-
Circuitry Linking the Catabolite Repression and Csr Global Regulatory Systems of Escherichia coli.J Bacteriol. 2016 Oct 7;198(21):3000-3015. doi: 10.1128/JB.00454-16. Print 2016 Nov 1. J Bacteriol. 2016. PMID: 27551019 Free PMC article.
-
Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB.Mol Microbiol. 1998 Sep;29(6):1321-30. doi: 10.1046/j.1365-2958.1998.01021.x. Mol Microbiol. 1998. PMID: 9781871 Review.
-
Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli.J Bacteriol. 2002 Sep;184(18):5130-40. doi: 10.1128/JB.184.18.5130-5140.2002. J Bacteriol. 2002. PMID: 12193630 Free PMC article.
-
The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli.J Biol Chem. 1997 Jul 11;272(28):17502-10. doi: 10.1074/jbc.272.28.17502. J Biol Chem. 1997. PMID: 9211896
-
CsrB sRNA family: sequestration of RNA-binding regulatory proteins.Curr Opin Microbiol. 2007 Apr;10(2):156-63. doi: 10.1016/j.mib.2007.03.007. Epub 2007 Mar 23. Curr Opin Microbiol. 2007. PMID: 17383221 Review.
Cited by
-
The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila.Infect Immun. 2004 Jun;72(6):3284-93. doi: 10.1128/IAI.72.6.3284-3293.2004. Infect Immun. 2004. PMID: 15155631 Free PMC article.
-
CsrA Shows Selective Regulation of sRNA-mRNA Networks.bioRxiv [Preprint]. 2023 Mar 29:2023.03.29.534774. doi: 10.1101/2023.03.29.534774. bioRxiv. 2023. PMID: 37034808 Free PMC article. Preprint.
-
Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium.Environ Microbiol. 2010 Feb;12(2):524-40. doi: 10.1111/j.1462-2920.2009.02097.x. Epub 2009 Nov 17. Environ Microbiol. 2010. PMID: 19919539 Free PMC article.
-
ppGpp: magic beyond RNA polymerase.Nat Rev Microbiol. 2012 Feb 16;10(3):203-12. doi: 10.1038/nrmicro2720. Nat Rev Microbiol. 2012. PMID: 22337166 Review.
-
CsrA Regulates Swarming Motility and Carbohydrate and Amino Acid Metabolism in Vibrio alginolyticus.Microorganisms. 2021 Nov 18;9(11):2383. doi: 10.3390/microorganisms9112383. Microorganisms. 2021. PMID: 34835507 Free PMC article.
References
-
- Altier C, Suyemoto M, Ruiz A I, Burnham K D, Maurer R. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol. 2000;35:635–646. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1989.
-
- Babitzke P. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol. 1997;26:1–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

