Counting the uncountable: statistical approaches to estimating microbial diversity
- PMID: 11571135
- PMCID: PMC93182
- DOI: 10.1128/AEM.67.10.4399-4406.2001
Counting the uncountable: statistical approaches to estimating microbial diversity
Erratum in
- Appl Environ Microbiol 2002 Jan;68(1):448
Figures

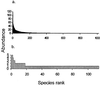
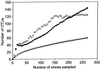
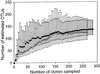
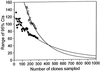
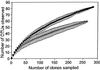

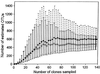
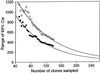
References
-
- Achenbach L A, Coates J D. Disparity between bacterial phylogeny and physiology. ASM News. 2000;66:714–715.
-
- Baltanas A. On the use of some methods for the estimation of species richness. Oikos. 1992;65:484–492.
-
- Bills G F, Polishook J D. Abundance and diversity of microfungi in leaf-litter of a lowland rain-forest in Costa Rica. Mycologia. 1994;86:187–198.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

