Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed
- PMID: 11571180
- PMCID: PMC93227
- DOI: 10.1128/AEM.67.10.4742-4751.2001
Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed
Abstract
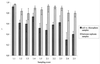
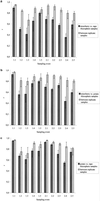
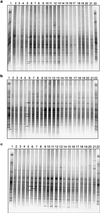
The bacterial rhizosphere communities of three host plants of the pathogenic fungus Verticillium dahliae, field-grown strawberry (Fragaria ananassa Duch.), oilseed rape (Brassica napus L.), and potato (Solanum tuberosum L.), were analyzed. We aimed to determine the degree to which the rhizosphere effect is plant dependent and whether this effect would be increased by growing the same crops in two consecutive years. Rhizosphere or soil samples were taken five times over the vegetation periods. To allow a cultivation-independent analysis, total community DNA was extracted from the microbial pellet recovered from root or soil samples. 16S rDNA fragments amplified by PCR from soil or rhizosphere bacterium DNA were analyzed by denaturing gradient gel electrophoresis (DGGE). The DGGE fingerprints showed plant-dependent shifts in the relative abundance of bacterial populations in the rhizosphere which became more pronounced in the second year. DGGE patterns of oilseed rape and potato rhizosphere communities were more similar to each other than to the strawberry patterns. In both years seasonal shifts in the abundance and composition of the bacterial rhizosphere populations were observed. Independent of the plant species, the patterns of the first sampling times for both years were characterized by the absence of some of the bands which became dominant at the following sampling times. Bacillus megaterium and Arthrobacter sp. were found as predominant populations in bulk soils. Sequencing of dominant bands excised from the rhizosphere patterns revealed that 6 out of 10 bands resembled gram-positive bacteria. Nocardia populations were identified as strawberry-specific bands.
Figures


References
-
- Bakken L R, Lindahl V. Recovery of bacterial cells from soil. In: Trevors J T, van Elsas J D, editors. Nucleic acids in the environment. Berlin, Germany: Springer-Verlag; 1995. pp. 9–27.
-
- Berg G, Kurze S, Buchner A, Wellington E M H, Smalla K. Successful strategy for the selection of new strawberry associated rhizobacteria antagonistic to Verticillium wilt. Can J Microbiol. 2000;46:1128–1137. - PubMed
-
- Berg, G., A. Fritze, N. Roskot, and K. Smalla. Evaluation of potential biocontrol rhizobacteria from different host plants of Verticillium dahliae Kleb. J. Appl. Microbiol., in press. - PubMed
-
- Brim H, Heuer H, Krögerrecklenfort E, Mergeay M, Smalla K. Characterization of the bacterial community of a zinc-polluted soil. Can J Microbiol. 1999;45:326–338. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

