Calcium regulation of neuronal gene expression
- PMID: 11572963
- PMCID: PMC58677
- DOI: 10.1073/pnas.191352298
Calcium regulation of neuronal gene expression
Abstract
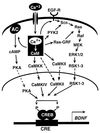
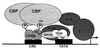
Plasticity is a remarkable feature of the brain, allowing neuronal structure and function to accommodate to patterns of electrical activity. One component of these long-term changes is the activity-driven induction of new gene expression, which is required for both the long-lasting long-term potentiation of synaptic transmission associated with learning and memory, and the activity dependent survival events that help to shape and wire the brain during development. We have characterized molecular mechanisms by which neuronal membrane depolarization and subsequent calcium influx into the cytoplasm lead to the induction of new gene transcription. We have identified three points within this cascade of events where the specificity of genes induced by different types of stimuli can be regulated. By using the induction of the gene that encodes brain-derived neurotrophic factor (BDNF) as a model, we have found that the ability of a calcium influx to induce transcription of this gene is regulated by the route of calcium entry into the cell, by the pattern of phosphorylation induced on the transcription factor cAMP-response element (CRE) binding protein (CREB), and by the complement of active transcription factors recruited to the BDNF promoter. These results refine and expand the working model of activity-induced gene induction in the brain, and help to explain how different types of neuronal stimuli can activate distinct transcriptional responses.
Figures


References
-
- Ghosh A, Greenberg M E. Science. 1995;268:239–247. - PubMed
-
- Franklin J L, Johnson E M. Trends Neurosci. 1992;15:501–508. - PubMed
-
- Lynch G, Larson J, Kelso S, Barrionuevo G, Schottler F. Nature (London) 1983;305:716–721. - PubMed
-
- Wu L, Wells D, Tay J, Mendis D, Abbott M A, Barnitt A, Quinlan E, Heynen A, Fallon J R, Richter J D. Neuron. 1998;21:1129–3119. - PubMed
-
- Aakalu G, Smith W B, Nguyen N, Jiang C, Schuman E M. Neuron. 2001;30:489–502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

