On the species of origin: diagnosing the source of symbiotic transcripts
- PMID: 11574056
- PMCID: PMC56898
- DOI: 10.1186/gb-2001-2-9-research0037
On the species of origin: diagnosing the source of symbiotic transcripts
Abstract
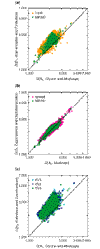
Background: Most organisms have developed ways to recognize and interact with other species. Symbiotic interactions range from pathogenic to mutualistic. Some molecular mechanisms of interspecific interaction are well understood, but many remain to be discovered. Expressed sequence tags (ESTs) from cultures of interacting symbionts can help identify transcripts that regulate symbiosis, but present a unique challenge for functional analysis. Given a sequence expressed in an interaction between two symbionts, the challenge is to determine from which organism the transcript originated. For high-throughput sequencing from interaction cultures, a reliable computational approach is needed. Previous investigations into GC nucleotide content and comparative similarity searching provide provisional solutions, but a comparative lexical analysis, which uses a likelihood-ratio test of hexamer counts, is more powerful.
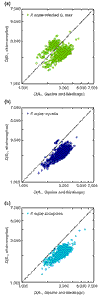
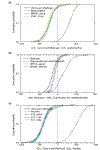
Results: Validation with genes whose origin and function are known yielded 94% accuracy. Microbial (non-plant) transcripts comprised 75% of a Phytophthora sojae-infected soybean (Glycine max cv Harasoy) library, contrasted with 15% or less in root tissue libraries of Medicago truncatula from axenic, Phytophthora medicaginis-infected, mycorrhizal, and rhizobacterial treatments. Mycorrhizal libraries contained about 23% microbial transcripts; an axenic plant library contained a similar proportion of putative microbial transcripts.
Conclusions: Comparative lexical analysis offers numerous advantages over alternative approaches. Many of the transcripts isolated from mixed cultures were of unknown function, suggesting specificity to symbiotic metabolism and therefore candidates likely to be interesting for further functional investigation. Future investigations will determine whether the abundance of non-plant transcripts in a pure plant library indicates procedural artifacts, horizontally transferred genes, or other phenomena.
Figures

References
-
- Adams MD, Fields C, Venter JC. Automated DNA sequencing and analysis London: Academic Press, 1994. - PubMed
-
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
-
- Cook DR. Medicago truncatula - a model in the making! Curr Opin Plant Biol. 1999;2:301–304. - PubMed
-
- Rounsley S, Lin X, Ketchum KA. Large-scale sequencing of plant genomes. Curr Opin Plant Biol. 1998;1:136–141. - PubMed
-
- Somerville C, Somerville S. Plant functional genomics. Science. 1999;285:380–383. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

