Design and development of a catalytic ribonucleoprotein
- PMID: 11574477
- PMCID: PMC125660
- DOI: 10.1093/emboj/20.19.5453
Design and development of a catalytic ribonucleoprotein
Abstract
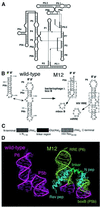
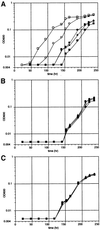
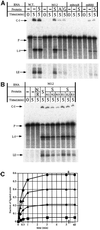
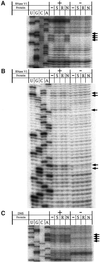
Ribonucleoproteins (RNPs) consisting of derivatives of a ribozyme and an RNA-binding protein were designed and constructed based upon high-resolution structures of the corresponding prototype molecules, the Tetrahymena group I self-splicing intron RNA and two proteins (bacteriophage lambdaN and HIV Rev proteins) containing RNA-binding motifs. The splicing reaction proceeds efficiently only when the designed RNA associates with the designed protein either in vivo or in vitro. In vivo mutagenic protein selection was effective for improving the capability of the protein. Kinetic analyses indicate that the protein promotes RNA folding to establish an active conformation. The fact that the conversion of a ribozyme to an RNP can be accomplished by simple molecular design supports the RNA world hypothesis and suggests that a natural active RNP might have evolved readily from a ribozyme.
Figures



References
-
- Battiste J.L., Mao,H., Rao,N.S., Tan,R., Muhandiram,D.R., Kay,L.E., Frankel,A.D. and Williamson,J.R. (1996) α helix–RNA major groove recognition in an HIV-1 rev peptide–RRE RNA complex. Science, 273, 1547–1551. - PubMed
-
- Burke J.M. (1996) Structural analysis and modifications of the hairpin ribozyme. In Eckstein,F. and Lilley,D.M.J. (eds), Catalytic RNA. Springer, Berlin, Germany, pp. 129–143.
-
- Caprara M.G., Mohr,G. and Lambowitz,A.M. (1996) A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol., 257, 512–531. - PubMed
-
- Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996a) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

