Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units
- PMID: 11574573
- PMCID: PMC88389
- DOI: 10.1128/JCM.39.10.3563-3571.2001
Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units
Abstract
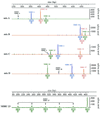
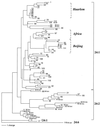
Large-scale genotyping of Mycobacterium tuberculosis is especially challenging, as the current typing methods are labor-intensive and the results are difficult to compare among laboratories. Here, automated typing based on variable-number tandem repeats (VNTRs) of genetic elements named mycobacterial interspersed repetitive units (MIRUs) in 12 mammalian minisatellite-like loci of M. tuberculosis is presented. This system combines analysis of multiplex PCRs on a fluorescence-based DNA analyzer with computerized automation of the genotyping. Analysis of a blinded reference set of 90 strains from 38 countries (K. Kremer et al., J. Clin. Microbiol. 37:2607-2618, 1999) demonstrated that it is 100% reproducible, sensitive, and specific for M. tuberculosis complex isolates, a performance that has not been achieved by any other typing method tested in the same conditions. MIRU-VNTRs can be used for analysis of the global genetic diversity of M. tuberculosis complex strains at different levels of evolutionary divergence. To fully exploit the portability of this typing system, a website was set up for the analysis of M. tuberculosis MIRU-VNTR genotypes via the Internet. This opens the way for global epidemiological surveillance of tuberculosis and should lead to novel insights into the evolutionary and population genetics of this major pathogen.
Figures
References
-
- Bifani P J, Mathema B, Liu Z, Moghazeh S L, Shopsin B, Tempalski B, Driscol J, Frothingham R, Musser J M, Alcabes P, Kreiswirth B N. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA. 1999;282:2321–2327. - PubMed
-
- Bifani P J, Plikaytis B B, Kapur V, Stockbauer K, Pan X, Lutfey M L, Moghazeh S L, Eisner W, Daniel T M, Kaplan M H, Crawford J T, Musser J M, Kreiswirth B N. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA. 1996;275:452–457. - PubMed
-
- Brosch R, Gordon S V, Eiglmeier K, Garnier T, Tekaia F, Yeramanian E, Cole S T. Genomics, biology, and evolution of the Mycobacterium tuberculosis complex. In: Hatful G F, Jacobs W R Jr, editors. Molecular genetics of mycobacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 19–36.
-
- Collins D M. Molecular epidemiology: Mycobacterium bovis. In: Ratledge C, Dale J, editors. Mycobacteria. Molecular biology and virulence. Oxford, United Kingdom: Blackwell Science Ltd.; 1999. pp. 123–135.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

