Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species
- PMID: 11574585
- PMCID: PMC88401
- DOI: 10.1128/JCM.39.10.3638-3648.2001
Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species
Erratum in
- J Clin Microbiol 2002 Jun;40(6):2316
Abstract
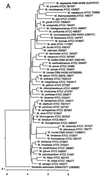
The use of the 16S rRNA gene for identification of nontuberculous mycobacteria (NTM) provides a faster and better ability to accurately identify them in addition to contributing significantly in the discovery of new species. Despite their associated problems, many rely on the use of public sequence databases for sequence comparisons. To best evaluate the taxonomic status of NTM species submitted to our reference laboratory, we have created a 16S rRNA sequence database by sequencing 121 American Type Culture Collection strains encompassing 92 species of mycobacteria, and have also included chosen unique mycobacterial sequences from public sequence repositories. In addition, the Ribosomal Differentiation of Medical Microorganisms (RIDOM) service has made freely available on the Internet mycobacterial identification by 16S rRNA analysis. We have evaluated 122 clinical NTM species using our database, comparing >1,400 bp of the 16S gene, and the RIDOM database, comparing approximately 440 bp. The breakdown of analysis was as follows: 61 strains had a sequence with 100% similarity to the type strain of an established species, 19 strains showed a 1- to 5-bp divergence from an established species, 11 strains had sequences corresponding to uncharacterized strain sequences in public databases, and 31 strains represented unique sequences. Our experience with analysis of the 16S rRNA gene of patient strains has shown that clear-cut results are not the rule. As many clinical, research, and environmental laboratories currently employ 16S-based identification of bacteria, including mycobacteria, a freely available quality-controlled database such as that provided by RIDOM is essential to accurately identify species or detect true sequence variations leading to the discovery of new species.
Figures

References
-
- Bercovier H, Kafri O, Sela S. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun. 1986;136:1136–1141. - PubMed
-
- Bottger E C, Kirschner P, Springer B, Zumft W. Mycobacteria degrading polycyclic aromatic hydrocarbons. Int J Syst Bacteriol. 1997;47:247.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

