Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin
- PMID: 11574687
- PMCID: PMC60231
- DOI: 10.1093/nar/29.19.4043
Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin
Abstract
In virtually all eukaryotic organisms, linker DNA between nucleosomes is associated with a histone termed linker histone or histone H1. In Saccharomyces cerevisiae, HHO1 encodes a putative linker histone with very significant homology to histone H1. The encoded protein is expressed in the nucleus, but has not been shown to affect global chromatin structure, nor has its deletion shown any detectable phenotype. In vitro chromatin assembly experiments with recombinant HHO1p have shown that it is able to complex with dinuncleosomes in a similar manner to histone H1. Here we report that while disruption of HHO1 has little affect on RNA levels of most cellular transcripts, there are numerous exceptions. Measurement of HHO1p concentration in the wild-type cell showed a stoichiometry of about one HHO1p molecule per 37 nucleosomes. Localization of HHO1p in the chromatin, using an immunoprecipitation technique, showed preferential HHO1p binding to rDNA sequences. These results suggest that HHO1p may play a similar role to linker histones, but at restricted locations in the chromatin.
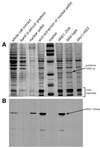
Figures



References
-
- Van Holde K.E. (1989) Chromatin. Springer-Verlag, New York, NY.
-
- Kornberg R.D. (1974) Chromatin structure: a repeating unit of histones and DNA. Science, 184, 868–871. - PubMed
-
- Kornberg R.D. and Thomas,J.O. (1974) Chromatin structure; oligomers of the histones. Science, 184, 865–868. - PubMed
-
- Graziano V., Gerchman,S.E., Schneider,D.K. and Ramakrishnan,V. (1994) Histone H1 is located in the interior of the chromatin 30-nm filament. Nature, 368, 351–354. - PubMed
-
- Doenecke D., Tonjes,R. and Kress,H. (1988) The H1 and core histone subtypes: differential gene expression and varied primary structures. Adv. Enzyme Regul., 27, 107–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

