Sugar additives for MALDI matrices improve signal allowing the smallest nucleotide change (A:T) in a DNA sequence to be resolved
- PMID: 11574693
- PMCID: PMC60251
- DOI: 10.1093/nar/29.19.e91
Sugar additives for MALDI matrices improve signal allowing the smallest nucleotide change (A:T) in a DNA sequence to be resolved
Abstract
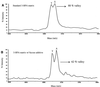
Sample preparation for matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS) of DNA is critical for obtaining high quality mass spectra. Sample impurity, solvent content, substrate surface and environmental conditions (temperature and humidity) all affect the rate of matrix-analyte co-crystallization. As a result, laser fluence threshold for desorption/ionization varies from spot to spot. When using 3-hydroxypicolinic acid (3-HPA) as the matrix, laser fluence higher than the threshold value reduces mass resolution in time-of-flight (TOF) MS as the excess energy transferred to DNA causes metastable decay. This can be overcome by either searching for 'hot' spots or adjusting the laser fluence. However, both solutions may require a significant amount of operator manipulation and are not ideal for automatic measurements. We have added various sugars for crystallization with the matrix to minimize the transfer of excess laser energy to DNA molecules. Fructose and fucose were found to be the most effective matrix additives. Using these additives, mass resolution for DNA molecules does not show noticeable deterioration as laser energy increases. Improved sample preparation is important for the detection of single nucleotide polymorphisms (SNPs) using primer extension with a single nucleotide. During automatic data acquisition it is difficult to routinely detect heterozygous A/T mutations, which requires resolving a mass difference of 9 Da, unless a sugar is added during crystallization.
Figures





References
-
- Ch’ang L.-Y., Tang,K., Schell,M., Ringelberg,C., Matteson,K.J., Allman,S.L. and Chen,C.H. (1995) Detection of ΔF508 mutation of the cystic fibrosis gene by matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom., 9, 772–774. - PubMed
-
- Bai J., Liu,Y., Liang,X., Zhu,Y. and Lubman,D.M. (1995) Procedures for detection of DNA by matrix-assisted laser desorption ionization mass spectrometry using a modified nafion film substrate. Rapid Commun. Mass Spectrom., 9, 1172–1176.
-
- Ross P.L. and Belgrader,P. (1997) Analysis of short tandem repeat polymorphisms in human DNA by matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem., 69, 3966–3972. - PubMed
-
- Ross P.L., Davis,P.A. and Belgrader,P. (1998) Analysis of DNA fragments from conventional and microfabricated PCR devices using delayed extraction MALDI-TOF mass spectrometry. Anal. Chem., 70, 2067–2073. - PubMed
-
- Taranenko N.I., Golovlev,V.V., Allman,S.L., Taranenko,N.V., Chen,C.H., Hong,J. and Chang,L.Y. (1998) Matrix-assisted laser desorption/ionization for short tandem repeat loci. Rapid Commun. Mass Spectrom., 12, 413–418. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

