Quantitative detection of single nucleotide polymorphisms for a pooled sample by a bioluminometric assay coupled with modified primer extension reactions (BAMPER)
- PMID: 11574695
- PMCID: PMC60253
- DOI: 10.1093/nar/29.19.e93
Quantitative detection of single nucleotide polymorphisms for a pooled sample by a bioluminometric assay coupled with modified primer extension reactions (BAMPER)
Abstract
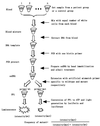
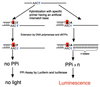
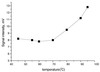
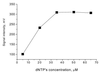
A new method for SNP analysis based on the detection of pyrophosphate (PPi) is demonstrated, which is capable of detecting small allele frequency differences between two DNA pools for genetic association studies other than SNP typing. The method is based on specific primer extension reactions coupled with PPi detection. As the specificity of the primer-directed extension is not enough for quantitative SNP analysis, artificial mismatched bases are introduced into the 3'-terminal regions of the specific primers as a way of improving the switching characteristics of the primer extension reactions. The best position in the primer for such artificial mismatched bases is the third position from the primer 3'-terminus. Contamination with endogenous PPi, which produces a large background signal level in SNP analysis, was removed using PPase to degrade the PPi during the sample preparation process. It is possible to accurately and quantitatively analyze SNPs using a set of primers that correspond to the wild-type and mutant DNA segments. The termini of these primers are at the mutation positions. Various types of SNPs were successfully analyzed. It was possible to very accurately determine SNPs with frequencies as low 0.02. It is very reproducible and the allele frequency difference can be determined. It is accurate enough to detect meaningful genetic differences among pooled DNA samples. The method is sensitive enough to detect 14 amol ssM13 DNA. The proposed method seems very promising in terms of realizing a cost-effective, large-scale human genetic testing system.
Figures




Similar articles
-
A gel-free SNP genotyping method: bioluminometric assay coupled with modified primer extension reactions (BAMPER) directly from double-stranded PCR products.Hum Mutat. 2004 Aug;24(2):155-63. doi: 10.1002/humu.20052. Hum Mutat. 2004. PMID: 15241797
-
Single-nucleotide polymorphism typing based on pyrosequencing chemistry and acryl-modified glass chip.Electrophoresis. 2009 Mar;30(6):991-8. doi: 10.1002/elps.200800395. Electrophoresis. 2009. PMID: 19235806
-
Single nucleotide polymorphism analysis by allele-specific primer extension with real-time bioluminescence detection in a microfluidic device.J Chromatogr A. 2003 Oct 3;1014(1-2):37-45. doi: 10.1016/s0021-9673(03)01033-1. J Chromatogr A. 2003. PMID: 14558610
-
Detection of single-base changes using a bioluminometric primer extension assay.Anal Biochem. 1997 Jan 15;244(2):367-73. doi: 10.1006/abio.1996.9913. Anal Biochem. 1997. PMID: 9025954
-
From gels to chips: "minisequencing" primer extension for analysis of point mutations and single nucleotide polymorphisms.Hum Mutat. 1999;13(1):1-10. doi: 10.1002/(SICI)1098-1004(1999)13:1<1::AID-HUMU1>3.0.CO;2-I. Hum Mutat. 1999. PMID: 9888384 Review.
Cited by
-
Determination of detection and quantification limits for SNP allele frequency estimation in DNA pools using real time PCR.Nucleic Acids Res. 2004 Feb 11;32(3):e24. doi: 10.1093/nar/gnh020. Nucleic Acids Res. 2004. PMID: 14960708 Free PMC article.
-
DNA analysis by fluorescence quenching detection.Genome Res. 2003 May;13(5):932-9. doi: 10.1101/gr.987803. Genome Res. 2003. PMID: 12727909 Free PMC article.
-
A new approach to SNP genotyping with fluorescently labeled mononucleotides.Nucleic Acids Res. 2004 Apr 15;32(7):e60. doi: 10.1093/nar/gnh054. Nucleic Acids Res. 2004. PMID: 15087492 Free PMC article.
-
Protocol: a simple gel-free method for SNP genotyping using allele-specific primers in rice and other plant species.Plant Methods. 2010 Apr 21;6:12. doi: 10.1186/1746-4811-6-12. Plant Methods. 2010. PMID: 20409329 Free PMC article.
-
Colorimetry-Based Phosphate Measurement for Polymerase Elongation.Biomed Res Int. 2023 Jan 23;2023:8296847. doi: 10.1155/2023/8296847. eCollection 2023. Biomed Res Int. 2023. PMID: 36726843 Free PMC article.
References
-
- Wang D.G., Fan,J.B., Siao,C.J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J. et al. (1998) Large-scale identification, mapping and genotyping of single-nucleotide polymorphisms in the human genome. Science, 280, 1077–1082. - PubMed
-
- See D., Kanazin,V., Talbert,H. and Blake,T. (2000) Electrophoretic detection of single-nucleotide polymorphisms. Biotechniques, 28, 710–714, 716. - PubMed
-
- Lyamichev V., Mast,A.L., Hall,J.G., Prudent,J.R., Kaiser,M.W., Takova,T., Kwiatkowski,R.W., Sander,T.J., de Arruda,M., Arco,D.A., Neri,B.P. and Brow,M.A. (1999) Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol., 17, 292–296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

