Chloroplast DNA methylation and inheritance in Chlamydomonas
- PMID: 11581163
- PMCID: PMC312798
- DOI: 10.1101/gad.906701
Chloroplast DNA methylation and inheritance in Chlamydomonas
Abstract
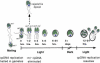
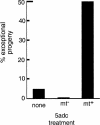
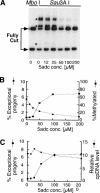
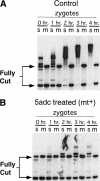
When Chlamydomonas reinhardtii cells mate, a zygotic maturation program is activated, part of which leads to destruction of chloroplast DNA (cpDNA) from the mating type minus (mt-) parent, and, therefore, to uniparental inheritance of mating type plus (mt+) cpDNA. A long-standing model that explains the selective destruction of mt(-) cpDNA in zygotes invokes a methylation-restriction system. We tested this model by using the potent methylation inhibitor 5-aza-2'-deoxycytidine (5adc) to hypomethylate parental cpDNA and found that the pattern of cpDNA inheritance is altered by 5adc in a manner that is consistent with the model. Surprisingly, however, hypomethylated mt+ cpDNA is not destroyed in zygotes as the methylation-restriction model predicts it should be. Destruction of mt- cpDNA is also unaffected when the parental mt+ cpDNA is hypomethylated. Instead, loss of methylation affects the relative rates of replication of residual mt- cpDNA and mt+ cpDNA in germinating zygotes. The mode of action for 5adc on cpDNA replication in germinating zygotes may be via hypomethylation of mt+ cpDNA, but is also consistent with its action as a DNA-damaging agent. Interestingly, 5adc causes reduced cpDNA replication only in germinating zygotes, not in vegetatively grown cells, indicating that cpDNA replication is qualitatively different in these two stages of the life cycle. Our results demonstrate that methylation is not necessary for protection of the mt+ cpDNA in early zygotes and uncover a novel stage of the Chlamydomonas life cycle when replication of cpDNA is highly susceptible to perturbation. Our data support a model in which differential cpDNA replication in germinating zygotes is used as a mechanism to selectively amplify intact and properly methylated cpDNA molecules.
Figures


References
-
- Ahmad I, Rao DN. Chemistry and biology of DNA methyltransferases. Crit Rev Biochem Mol Biol. 1996;31:361–380. - PubMed
-
- Armbrust EV. (eds. J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant). Kluwer Academic Pub, Dordrecht, The Netherlands. 1998. Uniparental inheritance of chloroplast genomes. In The molecular biology of chloroplasts and mitochondria in Chlamydomonas.
-
- Birky CW. Relaxed and stringent genomes: Why cytoplasmic genes don't obey Mendel's laws. J Hered. 1994;85:355–365.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
