Transgenic expression of survivin in keratinocytes counteracts UVB-induced apoptosis and cooperates with loss of p53
- PMID: 11581300
- PMCID: PMC200956
- DOI: 10.1172/JCI13345
Transgenic expression of survivin in keratinocytes counteracts UVB-induced apoptosis and cooperates with loss of p53
Abstract
The inhibitor of apoptosis protein survivin has been implicated in both cell cycle control and apoptosis resistance. To discriminate between these different roles, we used transgenic expression of survivin in the skin as a model for cell proliferation, differentiation, and apoptosis. Transgenic mice expressing survivin under the control of a keratin-14 promoter developed normally, without histologic abnormalities of the skin or hair, epidermal hyperplasia, or developmental abnormalities of basal or suprabasal epidermis. Keratinocyte proliferation assessed under basal conditions, or after ultraviolet-B (UVB) irradiation, or phorbol ester stimulation was unchanged in survivin transgenic mice. In contrast, survivin expression inhibited UVB-induced apoptosis in vitro and in vivo (i.e., sunburn cell formation), whereas it did not affect Fas-induced cell death. When crossed with p53 knockout mice, transgenic expression of survivin in a p53(+/-) background substituted for the loss of a second p53 allele and further inhibited UVB-induced apoptosis. These data provide the first in vivo evidence that survivin inhibits apoptosis and suggest that this pathway may oppose the elimination of cancerous cells by p53.
Figures
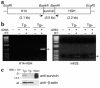
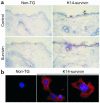



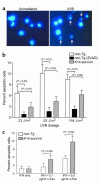

Comment in
-
The Survivin saga goes in vivo.J Clin Invest. 2001 Oct;108(7):965-9. doi: 10.1172/JCI14123. J Clin Invest. 2001. PMID: 11581297 Free PMC article. Review. No abstract available.
References
-
- Fuchs E. Keratins and the skin. Annu Rev Cell Dev Biol. 1995;11:123–153. - PubMed
-
- Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102:451–461. - PubMed
-
- Ishida-Yamamoto A, et al. Programmed cell death in normal epidermis and loricrin keratoderma. Multiple functions of profilaggrin in keratinization. J Investig Dermatol Symp Proc. 1999;4:145–149. - PubMed
-
- McCall CA, Cohen JJ. Programmed cell death in terminally differentiating keratinocytes: role of endogenous endonuclease. J Invest Dermatol. 1991;97:111–114. - PubMed
-
- Polakowska RR, Piacentini M, Bartlett R, Goldsmith LA, Haake AR. Apoptosis in human skin development: morphogenesis, periderm, and stem cells. Dev Dyn. 1994;199:176–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

