Analysis of thymocyte development reveals that the GTPase RhoA is a positive regulator of T cell receptor responses in vivo
- PMID: 11581313
- PMCID: PMC2193481
- DOI: 10.1084/jem.194.7.903
Analysis of thymocyte development reveals that the GTPase RhoA is a positive regulator of T cell receptor responses in vivo
Abstract
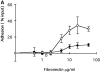
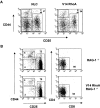
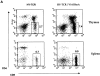
Loss of function of the guanine nucleotide binding protein RhoA blocks pre-T cell differentiation and survival indicating that this GTPase is a critical signaling molecule during early thymocyte development. Previous work has shown that the Rho family GTPase Rac-1 can initiate changes in actin dynamics necessary and sufficient for pre-T cell development. The present data now show that Rac-1 actions in pre-T cells require Rho function but that RhoA cannot substitute for Rac-1 and induce the actin cytoskeletal changes necessary for pre-T cell development. Activation of Rho is thus not sufficient to induce pre-T cell differentiation or survival in the absence of the pre-T cell receptor (TCR). The failure of RhoA activation to impact on pre-TCR-mediated signaling was in marked contrast to its actions on T cell responses mediated by the mature TCR alpha/beta complex. Cells expressing active RhoA were thus hyperresponsive in the context of TCR-induced proliferation in vitro and in vivo showed augmented positive selection of thymocytes expressing defined TCR complexes. This reveals that RhoA function is not only important for pre-T cells but also plays a role in determining the fate of mature T cells.
Figures












References
-
- Fehling H.J., von Boehmer H. Early αβ T cell development in the thymus of normal and genetically altered mice. Curr. Opin. Immunol. 1997;9:263–275. - PubMed
-
- von Boehmer H., Aifantis I., Feinberg J., Lechner O., Saint-Ruf C., Walter U., Buer J., Azogui O. Pleiotropic changes controlled by the pre-T-cell receptor. Curr. Opin. Immunol. 1999;11:135–142. - PubMed
-
- Jameson S.C., Bevan M.J. T-cell selection. Curr. Opin. Immunol. 1998;10:214–219. - PubMed
-
- Sebzda E., Mariathasan S., Ohteki T., Jones R., Bachmann M.F., Ohashi P.S. Selection of the T cell repertoire. Annu. Rev. Immunol. 1999;17:829–874. - PubMed
-
- Crespo P., Schuebel K.E., Ostrom A.A., Gutkind J.S., Bustelo X.R. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169–172. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

