mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor
- PMID: 11581395
- PMCID: PMC114601
- DOI: 10.1128/JVI.75.21.10272-10280.2001
mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor
Abstract
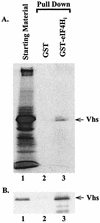
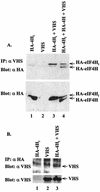
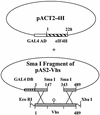
During lytic infections, the virion host shutoff (Vhs) protein (UL41) of herpes simplex virus destabilizes both host and viral mRNAs. By accelerating mRNA decay, it helps determine the levels and kinetics of viral and cellular gene expression. In vivo, Vhs shows a strong preference for mRNAs, as opposed to non-mRNAs, and degrades the 5' end of mRNAs prior to the 3' end. In contrast, partially purified Vhs is not restricted to mRNAs and causes cleavage of target RNAs at various sites throughout the molecule. To explain this discrepancy, we searched for cellular proteins that interact with Vhs using the Saccharomyces cerevisiae two-hybrid system. Vhs was found to interact with the human translation initiation factor, eIF4H. This interaction was verified by glutathione S-transferase pull-down experiments and by coimmunoprecipitation of Vhs and epitope-tagged eIF4H from extracts of mammalian cells. The interaction was abolished by several point mutations in Vhs that abrogate its ability to degrade mRNAs in vivo. The results suggest that Vhs is a viral mRNA degradation factor that is targeted to mRNAs, and to regions of translation initiation, through an interaction with eIF4H.
Figures



References
-
- Desai P, Person S. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology. 1996;220:516–521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

